Effect of Urban Wastewater Discharge on the Abundance of Antibiotic Resistance Genes and Antibiotic-Resistant Escherichia coli in Two Italian Rivers
- PMID: 32962009
- PMCID: PMC7557954
- DOI: 10.3390/ijerph17186813
Effect of Urban Wastewater Discharge on the Abundance of Antibiotic Resistance Genes and Antibiotic-Resistant Escherichia coli in Two Italian Rivers
Abstract
Background: Wastewater treatment plants (WWTPs) are microbial factories aimed to reduce the amount of nutrients and pathogenic microorganisms in the treated wastewater before its discharge into the environment. We studied the impact of urban WWTP effluents on the abundance of antibiotic resistance genes (ARGs) and antibiotic-resistant Escherichia coli (AR-E. coli) in the last stretch of two rivers (Arrone and Tiber) in Central Italy that differ in size and flow volume.
Methods: Water samples were collected in three seasons upstream and downstream of the WWTP, at the WWTP outlet, and at sea sites near the river mouth, and analyzed for the abundance of ARGs by qPCR and AR-E. coli using cultivation followed by disk diffusion assays.
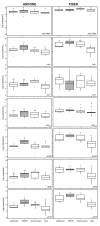
Results: For all studied genes (16S rRNA, intI1, sul1, ermB, blaTEM, tetW and qnrS), absolute concentrations were significantly higher in the Tiber than in the Arrone at all sampling sites, despite their collection date, but the prevalence of target ARGs within bacterial communities in both rivers was similar. The absolute concentrations of most ARGs were also generally higher in the WWTP effluent with median levels between log 4 and log 6 copies per ml but did not show differences along the studied stretches of rivers. Statistically significant site effect was found for E. coli phenotypic resistance to tetracycline and ciprofloxacin in the Arrone but not in the Tiber.
Conclusions: In both rivers, diffuse or point pollution sources other than the studied WWTP effluents may account for the observed resistance pattern, although the Arrone appears as more sensitive to the wastewater impact considering its lower flow volume.
Keywords: Arrone River; Escherichia coli; Tiber River; antibiotic resistance; antibiotic resistance genes.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

