Clearing of Foreign Episomal DNA from Human Cells by CRISPRa-Mediated Activation of Cytidine Deaminases
- PMID: 32962129
- PMCID: PMC7557733
- DOI: 10.3390/ijms21186865
Clearing of Foreign Episomal DNA from Human Cells by CRISPRa-Mediated Activation of Cytidine Deaminases
Abstract
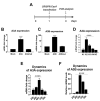
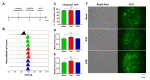
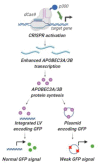
Restriction of foreign DNA is a fundamental defense mechanism required for maintaining genomic stability and proper function of mammalian cells. APOBEC cytidine deaminases are crucial effector molecules involved in clearing pathogenic DNA of viruses and other microorganisms and improperly localized self-DNA (DNA leakages). Mastering the expression of APOBEC provides the crucial means both for developing novel therapeutic approaches for combating infectious and non-infectious diseases and for numerous research purposes. In this study, we report successful application of a CRISPRa approach to effectively and specifically overexpress APOBEC3A and APOBEC3B deaminases and describe their effects on episomal and integrated foreign DNA. This method increased target gene transcription by >6-50-fold in HEK293T cells. Furthermore, CRISPRa-mediated activation of APOBEC3A/APOBEC3B suppressed episomal but not integrated foreign DNA. Episomal GC-rich DNA was rapidly destabilized and destroyed by CRISPRa-induced APOBEC3A/APOBEC3B, while the remaining DNA templates harbored frequent deaminated nucleotides. To conclude, the CRISPRa approach could be readily utilized for manipulating innate immunity and investigating the effects of the key effector molecules on foreign nucleic acids.
Keywords: APOBECs; CRISPRa; cytidine deaminases; deamination; foreign DNA; innate immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

