Influenza-Host Interplay and Strategies for Universal Vaccine Development
- PMID: 32962304
- PMCID: PMC7564814
- DOI: 10.3390/vaccines8030548
Influenza-Host Interplay and Strategies for Universal Vaccine Development
Abstract
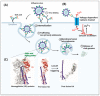
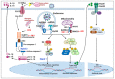
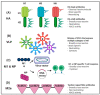
Influenza is an annual epidemic and an occasional pandemic caused by pathogens that are responsible for infectious respiratory disease. Humans are highly susceptible to the infection mediated by influenza A viruses (IAV). The entry of the virus is mediated by the influenza virus hemagglutinin (HA) glycoprotein that binds to the cellular sialic acid receptors and facilitates the fusion of the viral membrane with the endosomal membrane. During IAV infection, virus-derived pathogen-associated molecular patterns (PAMPs) are recognized by host intracellular specific sensors including toll-like receptors (TLRs), C-type lectin receptors, retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) either on the cell surface or intracellularly in endosomes. Herein, we comprehensively review the current knowledge available on the entry of the influenza virus into host cells and the molecular details of the influenza virus-host interface. We also highlight certain strategies for the development of universal influenza vaccines.
Keywords: adaptive immune response; immunopathology; influenza A virus; innate immune response; universal influenza vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Innate and intrinsic antiviral immunity in skin.J Dermatol Sci. 2014 Sep;75(3):159-66. doi: 10.1016/j.jdermsci.2014.05.004. Epub 2014 Jun 2. J Dermatol Sci. 2014. PMID: 24928148 Review.
-
RNA Sensors as a Mechanism of Innate Immune Evasion among SARSCoV2, HIV and Nipah Viruses.Curr Protein Pept Sci. 2021 Oct 26;22(4):273-289. doi: 10.2174/1389203722666210322142725. Curr Protein Pept Sci. 2021. PMID: 33749559 Review.
-
Mechanisms and pathways of innate immune activation and regulation in health and cancer.Hum Vaccin Immunother. 2014;10(11):3270-85. doi: 10.4161/21645515.2014.979640. Hum Vaccin Immunother. 2014. PMID: 25625930 Free PMC article. Review.
-
De Novo Transcriptome Analysis Shows That SAV-3 Infection Upregulates Pattern Recognition Receptors of the Endosomal Toll-Like and RIG-I-Like Receptor Signaling Pathways in Macrophage/Dendritic Like TO-Cells.Viruses. 2016 Apr 21;8(4):114. doi: 10.3390/v8040114. Viruses. 2016. PMID: 27110808 Free PMC article.
-
Casein Kinase 1α Mediates the Degradation of Receptors for Type I and Type II Interferons Caused by Hemagglutinin of Influenza A Virus.J Virol. 2018 Mar 14;92(7):e00006-18. doi: 10.1128/JVI.00006-18. Print 2018 Apr 1. J Virol. 2018. PMID: 29343571 Free PMC article.
Cited by
-
Influenza A (N1-N9) and Influenza B (B/Victoria and B/Yamagata) Neuraminidase Pseudotypes as Tools for Pandemic Preparedness and Improved Influenza Vaccine Design.Vaccines (Basel). 2022 Sep 14;10(9):1520. doi: 10.3390/vaccines10091520. Vaccines (Basel). 2022. PMID: 36146598 Free PMC article.
-
Endocytosis of abiotic nanomaterials and nanobiovectors: Inhibition of membrane trafficking.Nano Today. 2021 Oct;40:101279. doi: 10.1016/j.nantod.2021.101279. Epub 2021 Sep 8. Nano Today. 2021. PMID: 34518771 Free PMC article. Review.
-
The PB1 protein of H9N2 influenza A virus inhibits antiviral innate immunity by targeting MAVS for TRIM25-mediated autophagic degradation.Poult Sci. 2025 Jan;104(1):104639. doi: 10.1016/j.psj.2024.104639. Epub 2024 Dec 4. Poult Sci. 2025. PMID: 39647358 Free PMC article.
-
Transcriptional Analysis of lncRNA and Target Genes Induced by Influenza A Virus Infection in MDCK Cells.Vaccines (Basel). 2023 Oct 14;11(10):1593. doi: 10.3390/vaccines11101593. Vaccines (Basel). 2023. PMID: 37896995 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

