Outer Membrane Disruption Overcomes Intrinsic, Acquired, and Spontaneous Antibiotic Resistance
- PMID: 32963002
- PMCID: PMC7512548
- DOI: 10.1128/mBio.01615-20
Outer Membrane Disruption Overcomes Intrinsic, Acquired, and Spontaneous Antibiotic Resistance
Abstract
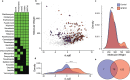
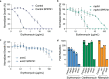
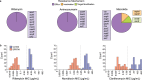
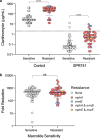
Disruption of the outer membrane (OM) barrier allows for the entry of otherwise inactive antimicrobials into Gram-negative pathogens. Numerous efforts to implement this approach have identified a large number of OM perturbants that sensitize Gram-negative bacteria to many clinically available Gram-positive active antibiotics. However, there is a dearth of investigation into the strengths and limitations of this therapeutic strategy, with an overwhelming focus on characterization of individual potentiator molecules. Herein, we look to explore the utility of exploiting OM perturbation to sensitize Gram-negative pathogens to otherwise inactive antimicrobials. We identify the ability of OM disruption to change the rules of Gram-negative entry, overcome preexisting and spontaneous resistance, and impact biofilm formation. Disruption of the OM expands the threshold of hydrophobicity compatible with Gram-negative activity to include hydrophobic molecules. We demonstrate that while resistance to Gram-positive active antibiotics is surprisingly common in Gram-negative pathogens, OM perturbation overcomes many antibiotic inactivation determinants. Further, we find that OM perturbation reduces the rate of spontaneous resistance to rifampicin and impairs biofilm formation. Together, these data suggest that OM disruption overcomes many of the traditional hurdles encountered during antibiotic treatment and is a high priority approach for further development.IMPORTANCE The spread of antibiotic resistance is an urgent threat to global health that necessitates new therapeutics. Treatments for Gram-negative pathogens are particularly challenging to identify due to the robust outer membrane permeability barrier in these organisms. Recent discovery efforts have attempted to overcome this hurdle by disrupting the outer membrane using chemical perturbants and have yielded several new peptides and small molecules that allow the entry of otherwise inactive antimicrobials. However, a comprehensive investigation into the strengths and limitations of outer membrane perturbants as antibiotic partners is currently lacking. Herein, we interrogate the interaction between outer membrane perturbation and several common impediments to effective antibiotic use. Interestingly, we discover that outer membrane disruption is able to overcome intrinsic, spontaneous, and acquired antibiotic resistance in Gram-negative bacteria, meriting increased attention toward this approach.
Keywords: Gram-negative; antibiotic adjuvant; antibiotic resistance; outer membrane.
Copyright © 2020 MacNair and Brown.
Figures
Similar articles
-
Scope and Limitations of Exploiting the Ability of the Chemosensitizer NV716 to Enhance the Activity of Tetracycline Derivatives against Pseudomonas aeruginosa.Molecules. 2023 May 23;28(11):4262. doi: 10.3390/molecules28114262. Molecules. 2023. PMID: 37298737 Free PMC article.
-
Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria.mBio. 2017 Oct 31;8(5):e01172-17. doi: 10.1128/mBio.01172-17. mBio. 2017. PMID: 29089426 Free PMC article.
-
New potentiators of ineffective antibiotics: Targeting the Gram-negative outer membrane to overcome intrinsic resistance.Curr Opin Chem Biol. 2022 Feb;66:102099. doi: 10.1016/j.cbpa.2021.102099. Epub 2021 Nov 19. Curr Opin Chem Biol. 2022. PMID: 34808425 Review.
-
IMT-P8 potentiates Gram-positive specific antibiotics in intrinsically resistant Gram-negative bacteria.Antimicrob Agents Chemother. 2024 Oct 8;68(10):e0075324. doi: 10.1128/aac.00753-24. Epub 2024 Sep 5. Antimicrob Agents Chemother. 2024. PMID: 39235250 Free PMC article.
-
Treatment of Gram-negative bacterial infections by potentiation of antibiotics.Curr Opin Microbiol. 2016 Oct;33:7-12. doi: 10.1016/j.mib.2016.05.005. Epub 2016 May 25. Curr Opin Microbiol. 2016. PMID: 27232956 Review.
Cited by
-
Synergistic Inhibition of Plantaricin E/F and Lactic Acid Against Aeromonas hydrophila LPL-1 Reveals the Novel Potential of Class IIb Bacteriocin.Front Microbiol. 2022 Feb 15;13:774184. doi: 10.3389/fmicb.2022.774184. eCollection 2022. Front Microbiol. 2022. PMID: 35242114 Free PMC article.
-
Role of AlgC and GalU in the Intrinsic Antibiotic Resistance of Helicobacter pylori.Infect Drug Resist. 2023 Mar 29;16:1839-1847. doi: 10.2147/IDR.S403046. eCollection 2023. Infect Drug Resist. 2023. PMID: 37016632 Free PMC article.
-
OmpH is Involved in the Decrease of Acinetobacter baumannii Biofilm by the Antimicrobial Peptide Cec4.Drug Des Devel Ther. 2024 Dec 7;18:5795-5810. doi: 10.2147/DDDT.S481225. eCollection 2024. Drug Des Devel Ther. 2024. PMID: 39664965 Free PMC article.
-
Novel Antibacterial Approaches and Therapeutic Strategies.Antibiotics (Basel). 2025 Apr 15;14(4):404. doi: 10.3390/antibiotics14040404. Antibiotics (Basel). 2025. PMID: 40298586 Free PMC article. Review.
-
Loss of β-Ketoacyl Acyl Carrier Protein Synthase III Activity Restores Multidrug-Resistant Escherichia coli Sensitivity to Previously Ineffective Antibiotics.mSphere. 2022 Jun 29;7(3):e0011722. doi: 10.1128/msphere.00117-22. Epub 2022 May 16. mSphere. 2022. PMID: 35574679 Free PMC article.
References
-
- Jernigan JA, Hatfield KM, Wolford H, Nelson RE, Olubajo B, Reddy SC, McCarthy N, Paul P, McDonald LC, Kallen A, Fiore A, Craig M, Baggs J. 2020. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med 382:1309–1319. doi:10.1056/NEJMoa1914433. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

