Distinct contributions of DNA methylation and histone acetylation to the genomic occupancy of transcription factors
- PMID: 32963030
- PMCID: PMC7605266
- DOI: 10.1101/gr.257576.119
Distinct contributions of DNA methylation and histone acetylation to the genomic occupancy of transcription factors
Abstract
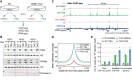
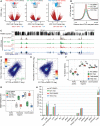
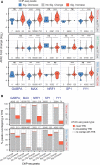
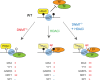
Epigenetic modifications on chromatin play important roles in regulating gene expression. Although chromatin states are often governed by multilayered structure, how individual pathways contribute to gene expression remains poorly understood. For example, DNA methylation is known to regulate transcription factor binding but also to recruit methyl-CpG binding proteins that affect chromatin structure through the activity of histone deacetylase complexes (HDACs). Both of these mechanisms can potentially affect gene expression, but the importance of each, and whether these activities are integrated to achieve appropriate gene regulation, remains largely unknown. To address this important question, we measured gene expression, chromatin accessibility, and transcription factor occupancy in wild-type or DNA methylation-deficient mouse embryonic stem cells following HDAC inhibition. We observe widespread increases in chromatin accessibility at retrotransposons when HDACs are inhibited, and this is magnified when cells also lack DNA methylation. A subset of these elements has elevated binding of the YY1 and GABPA transcription factors and increased expression. The pronounced additive effect of HDAC inhibition in DNA methylation-deficient cells demonstrates that DNA methylation and histone deacetylation act largely independently to suppress transcription factor binding and gene expression.
© 2020 Cusack et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
