Coupled intra- and interdomain dynamics support domain cross-talk in Pin1
- PMID: 32963105
- PMCID: PMC7864058
- DOI: 10.1074/jbc.RA120.015849
Coupled intra- and interdomain dynamics support domain cross-talk in Pin1
Abstract
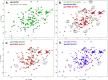
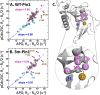
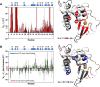
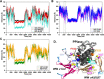
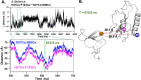
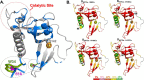
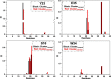
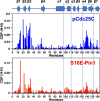
The functional mechanisms of multidomain proteins often exploit interdomain interactions, or "cross-talk." An example is human Pin1, an essential mitotic regulator consisting of a Trp-Trp (WW) domain flexibly tethered to a peptidyl-prolyl isomerase (PPIase) domain, resulting in interdomain interactions important for Pin1 function. Substrate binding to the WW domain alters its transient contacts with the PPIase domain via means that are only partially understood. Accordingly, we have investigated Pin1 interdomain interactions using NMR paramagnetic relaxation enhancement (PRE) and molecular dynamics (MD) simulations. The PREs show that apo-Pin1 samples interdomain contacts beyond the range suggested by previous structural studies. They further show that substrate binding to the WW domain simultaneously alters interdomain separation and the internal conformation of the WW domain. A 4.5-μs all-atom MD simulation of apo-Pin1 suggests that the fluctuations of interdomain distances are correlated with fluctuations of WW domain interresidue contacts involved in substrate binding. Thus, the interdomain/WW domain conformations sampled by apo-Pin1 may already include a range of conformations appropriate for binding Pin1's numerous substrates. The proposed coupling between intra-/interdomain conformational fluctuations is a consequence of the dynamic modular architecture of Pin1. Such modular architecture is common among cell-cycle proteins; thus, the WW-PPIase domain cross-talk mechanisms of Pin1 may be relevant for their mechanisms as well.
Keywords: MD; Pin1; allosteric regulation; multidomain protein; nuclear magnetic resonance (NMR); paramagnetic relaxation enhancement; post-translational modification; post-translational modification (PTM); protein dynamic; protein-protein interaction.
© 2020 Zhang et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Similar articles
-
Interdomain interactions support interdomain communication in human Pin1.Biochemistry. 2013 Oct 8;52(40):6968-81. doi: 10.1021/bi401057x. Epub 2013 Sep 24. Biochemistry. 2013. PMID: 24020391 Free PMC article.
-
Uncorrelated Effect of Interdomain Contact on Pin1 Isomerase Activity Reveals Positive Catalytic Cooperativity.J Phys Chem Lett. 2019 Mar 21;10(6):1272-1278. doi: 10.1021/acs.jpclett.9b00052. Epub 2019 Mar 6. J Phys Chem Lett. 2019. PMID: 30821977
-
Activity and Affinity of Pin1 Variants.Molecules. 2019 Dec 20;25(1):36. doi: 10.3390/molecules25010036. Molecules. 2019. PMID: 31861908 Free PMC article. Review.
-
Dynamic Allostery Modulates Catalytic Activity by Modifying the Hydrogen Bonding Network in the Catalytic Site of Human Pin1.Molecules. 2017 Jun 15;22(6):992. doi: 10.3390/molecules22060992. Molecules. 2017. PMID: 28617332 Free PMC article.
-
Gears-In-Motion: The Interplay of WW and PPIase Domains in Pin1.Front Oncol. 2018 Oct 25;8:469. doi: 10.3389/fonc.2018.00469. eCollection 2018. Front Oncol. 2018. PMID: 30460195 Free PMC article. Review.
Cited by
-
A tethering mechanism underlies Pin1-catalyzed proline cis-trans isomerization at a noncanonical site.Proc Natl Acad Sci U S A. 2025 May 27;122(21):e2414606122. doi: 10.1073/pnas.2414606122. Epub 2025 May 19. Proc Natl Acad Sci U S A. 2025. PMID: 40388619
-
A tethering mechanism underlies Pin1-catalyzed proline cis-trans isomerization at a noncanonical site.bioRxiv [Preprint]. 2025 Mar 22:2024.07.19.604348. doi: 10.1101/2024.07.19.604348. bioRxiv. 2025. Update in: Proc Natl Acad Sci U S A. 2025 May 27;122(21):e2414606122. doi: 10.1073/pnas.2414606122. PMID: 39091828 Free PMC article. Updated. Preprint.
-
Ligand-specific conformational change drives interdomain allostery in Pin1.Nat Commun. 2022 Aug 4;13(1):4546. doi: 10.1038/s41467-022-32340-x. Nat Commun. 2022. PMID: 35927276 Free PMC article.
-
Identification and inhibition of PIN1-NRF2 protein-protein interactions through computational and biophysical approaches.Sci Rep. 2025 Mar 14;15(1):8907. doi: 10.1038/s41598-025-89342-0. Sci Rep. 2025. PMID: 40087364 Free PMC article.
-
What's all the phos about? Insights into the phosphorylation state of the RNA polymerase II C-terminal domain via mass spectrometry.RSC Chem Biol. 2021 Jun 3;2(4):1084-1095. doi: 10.1039/d1cb00083g. eCollection 2021 Aug 5. RSC Chem Biol. 2021. PMID: 34458825 Free PMC article. Review.
References
-
- Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Counter C.M., Nevins J.R., Means A.R., Sears R. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. 15048125. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

