Genetic correlations and genome-wide associations of cortical structure in general population samples of 22,824 adults
- PMID: 32963231
- PMCID: PMC7508833
- DOI: 10.1038/s41467-020-18367-y
Genetic correlations and genome-wide associations of cortical structure in general population samples of 22,824 adults
Abstract
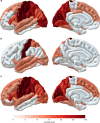
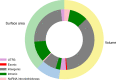
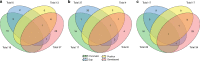
Cortical thickness, surface area and volumes vary with age and cognitive function, and in neurological and psychiatric diseases. Here we report heritability, genetic correlations and genome-wide associations of these cortical measures across the whole cortex, and in 34 anatomically predefined regions. Our discovery sample comprises 22,824 individuals from 20 cohorts within the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium and the UK Biobank. We identify genetic heterogeneity between cortical measures and brain regions, and 160 genome-wide significant associations pointing to wnt/β-catenin, TGF-β and sonic hedgehog pathways. There is enrichment for genes involved in anthropometric traits, hindbrain development, vascular and neurodegenerative disease and psychiatric conditions. These data are a rich resource for studies of the biological mechanisms behind cortical development and aging.
Conflict of interest statement
Dr. Dale is a founder of and holds equity in CorTechs Labs, Inc, and serves on its Scientific Advisory Board. He is a member of the Scientific Advisory Board of Human Longevity, Inc. and receives funding through research agreements with General Electric Healthcare and Medtronic, Inc. The terms of these arrangements have been reviewed and approved by UCSD in accordance with its conflict of interest policies. W. Niessen is co-founder and shareholder of Quantib BV. H. Brodaty is an advisory board member of Nutricia. The other authors declare no competing interests.
Figures

Similar articles
-
Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function.Nat Commun. 2018 May 29;9(1):2098. doi: 10.1038/s41467-018-04362-x. Nat Commun. 2018. PMID: 29844566 Free PMC article.
-
Identification of Genetic Loci Jointly Influencing Schizophrenia Risk and the Cognitive Traits of Verbal-Numerical Reasoning, Reaction Time, and General Cognitive Function.JAMA Psychiatry. 2017 Oct 1;74(10):1065-1075. doi: 10.1001/jamapsychiatry.2017.1986. JAMA Psychiatry. 2017. PMID: 28746715 Free PMC article.
-
The genomic basis of mood instability: identification of 46 loci in 363,705 UK Biobank participants, genetic correlation with psychiatric disorders, and association with gene expression and function.Mol Psychiatry. 2020 Nov;25(11):3091-3099. doi: 10.1038/s41380-019-0439-8. Epub 2019 Jun 5. Mol Psychiatry. 2020. PMID: 31168069 Free PMC article.
-
The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals.J Gerontol A Biol Sci Med Sci. 2012 May;67(5):470-9. doi: 10.1093/gerona/gls089. Epub 2012 Apr 12. J Gerontol A Biol Sci Med Sci. 2012. PMID: 22499766 Free PMC article. Review.
-
[Genomic instability in the brain: etiology, pathogenesis and new biological markers of psychiatric disorders].Vestn Ross Akad Med Nauk. 2012;(9):45-53. Vestn Ross Akad Med Nauk. 2012. PMID: 23210172 Review. Russian.
Cited by
-
Multivariate genome-wide association study on tissue-sensitive diffusion metrics highlights pathways that shape the human brain.Nat Commun. 2022 May 3;13(1):2423. doi: 10.1038/s41467-022-30110-3. Nat Commun. 2022. PMID: 35505052 Free PMC article.
-
From base pair to brain.Nat Neurosci. 2021 May;24(5):619-621. doi: 10.1038/s41593-021-00852-2. Nat Neurosci. 2021. PMID: 33875895 No abstract available.
-
Integrated Genomic, Transcriptomic and Proteomic Analysis for Identifying Markers of Alzheimer's Disease.Diagnostics (Basel). 2021 Dec 8;11(12):2303. doi: 10.3390/diagnostics11122303. Diagnostics (Basel). 2021. PMID: 34943540 Free PMC article.
-
Polygenic risk scores and brain structures both contribute to externalizing behavior in childhood - A study in the Adolescent Brain and Cognitive Development (ABCD) cohort.Neurosci Appl. 2023 May 24;2:101128. doi: 10.1016/j.nsa.2023.101128. eCollection 2023. Neurosci Appl. 2023. PMID: 40655980 Free PMC article.
-
Enhanced insights into the genetic architecture of 3D cranial vault shape using pleiotropy-informed GWAS.Commun Biol. 2025 Mar 15;8(1):439. doi: 10.1038/s42003-025-07875-6. Commun Biol. 2025. PMID: 40087503 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- N01HC55222/HL/NHLBI NIH HHS/United States
- N01HC25195/HL/NHLBI NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- N01HC85081/HL/NHLBI NIH HHS/United States
- R01 AG054076/AG/NIA NIH HHS/United States
- P41 EB015922/EB/NIBIB NIH HHS/United States
- N01HC85080/HL/NHLBI NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- MR/M01311/1/MRC_/Medical Research Council/United Kingdom
- G1001401/MRC_/Medical Research Council/United Kingdom
- TMH109788/CIHR/Canada
- BB/F019394/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 AG023629/AG/NIA NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01 EB015611/EB/NIBIB NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- R01 NS087541/NS/NINDS NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- R01 HL103612/HL/NHLBI NIH HHS/United States
- R01 AG049607/AG/NIA NIH HHS/United States
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- MR/M013111/1/MRC_/Medical Research Council/United Kingdom
- G1001245/MRC_/Medical Research Council/United Kingdom
- MR/R024065/1/MRC_/Medical Research Council/United Kingdom
- N01HC85082/HL/NHLBI NIH HHS/United States
- U54 EB020403/EB/NIBIB NIH HHS/United States
- NET54015/CIHR/Canada
- N02 HL64278/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- MR/N027558/1/MRC_/Medical Research Council/United Kingdom
- N01HC85079/HL/NHLBI NIH HHS/United States
- N01HC85083/HL/NHLBI NIH HHS/United States
- N01HC85086/HL/NHLBI NIH HHS/United States
- G0701120/MRC_/Medical Research Council/United Kingdom
- R01 AG033040/AG/NIA NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- R01 AG016495/AG/NIA NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- R01 MH117601/MH/NIMH NIH HHS/United States
- RF1 AG051710/AG/NIA NIH HHS/United States
- R01 AG059874/AG/NIA NIH HHS/United States
- 75N92019D00031/HL/NHLBI NIH HHS/United States
- G0700704/MRC_/Medical Research Council/United Kingdom
- R01 HL087652/HL/NHLBI NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- S10 OD023696/OD/NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- G0700704/84698/MRC_/Medical Research Council/United Kingdom
- NRF86678/CIHR/Canada
- R01 MH116147/MH/NIMH NIH HHS/United States
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- RF1 AG041915/AG/NIA NIH HHS/United States
- R01 AG022381/AG/NIA NIH HHS/United States
- R01 AG050595/AG/NIA NIH HHS/United States
- 8200/MRC_/Medical Research Council/United Kingdom
- P01 AG026572/AG/NIA NIH HHS/United States
- U01 AG049505/AG/NIA NIH HHS/United States

