A diploid assembly-based benchmark for variants in the major histocompatibility complex
- PMID: 32963235
- PMCID: PMC7508831
- DOI: 10.1038/s41467-020-18564-9
A diploid assembly-based benchmark for variants in the major histocompatibility complex
Abstract
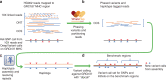
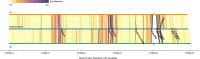
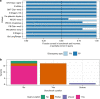
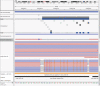
Most human genomes are characterized by aligning individual reads to the reference genome, but accurate long reads and linked reads now enable us to construct accurate, phased de novo assemblies. We focus on a medically important, highly variable, 5 million base-pair (bp) region where diploid assembly is particularly useful - the Major Histocompatibility Complex (MHC). Here, we develop a human genome benchmark derived from a diploid assembly for the openly-consented Genome in a Bottle sample HG002. We assemble a single contig for each haplotype, align them to the reference, call phased small and structural variants, and define a small variant benchmark for the MHC, covering 94% of the MHC and 22368 variants smaller than 50 bp, 49% more variants than a mapping-based benchmark. This benchmark reliably identifies errors in mapping-based callsets, and enables performance assessment in regions with much denser, complex variation than regions covered by previous benchmarks.
Conflict of interest statement
C.-S.C. and A.F. are employees of DNAnexus Inc., a company providing a cloud computing platform for processing genomic information. C.-S.C. is a co-founder and partner of Omni BioComputing, LLC, which currently develops genome assembler related technologies. Q.Z. is an employee of Laboratory Corporation of America Holdings, a company providing clinical diagnostics services. A.T.D. is a partner in Peptide Groove, LLP. A.C. is an employee of Google, a company providing a cloud computing platform. W.J.R. is an employee and shareholder of Pacific Biosciences. A.M.B. is an ex-employee and shareholder of 10x Genomics. The other authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

