Consensus transcriptional regulatory networks of coronavirus-infected human cells
- PMID: 32963239
- PMCID: PMC7509801
- DOI: 10.1038/s41597-020-00628-6
Consensus transcriptional regulatory networks of coronavirus-infected human cells
Abstract
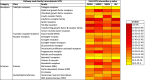
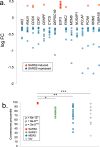
Establishing consensus around the transcriptional interface between coronavirus (CoV) infection and human cellular signaling pathways can catalyze the development of novel anti-CoV therapeutics. Here, we used publicly archived transcriptomic datasets to compute consensus regulatory signatures, or consensomes, that rank human genes based on their rates of differential expression in MERS-CoV (MERS), SARS-CoV-1 (SARS1) and SARS-CoV-2 (SARS2)-infected cells. Validating the CoV consensomes, we show that high confidence transcriptional targets (HCTs) of MERS, SARS1 and SARS2 infection intersect with HCTs of signaling pathway nodes with known roles in CoV infection. Among a series of novel use cases, we gather evidence for hypotheses that SARS2 infection efficiently represses E2F family HCTs encoding key drivers of DNA replication and the cell cycle; that progesterone receptor signaling antagonizes SARS2-induced inflammatory signaling in the airway epithelium; and that SARS2 HCTs are enriched for genes involved in epithelial to mesenchymal transition. The CoV infection consensomes and HCT intersection analyses are freely accessible through the Signaling Pathways Project knowledgebase, and as Cytoscape-style networks in the Network Data Exchange repository.
Conflict of interest statement
The authors declare no competing interests.
Figures







Update of
-
Consensus transcriptional regulatory networks of coronavirus-infected human cells.bioRxiv [Preprint]. 2020 Jul 15:2020.04.24.059527. doi: 10.1101/2020.04.24.059527. bioRxiv. 2020. Update in: Sci Data. 2020 Sep 22;7(1):314. doi: 10.1038/s41597-020-00628-6. PMID: 32511379 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

