MicroRNA-Related Prognosis Biomarkers from High-Throughput Sequencing Data of Colorectal Cancer
- PMID: 32964043
- PMCID: PMC7501550
- DOI: 10.1155/2020/7905380
MicroRNA-Related Prognosis Biomarkers from High-Throughput Sequencing Data of Colorectal Cancer
Abstract
Background: Colorectal cancer (CRC) is the third most common cancer in the world, and most of them are adenocarcinomas. CRC could be classified as colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) according to the original tumorigenesis position. Increasing evidences indicated that microRNAs (miRNAs) play an important role in the occurrence of multiple tumors.
Methods: In this study, we firstly downloaded miRNA (COAD, 8 controls vs. 455 tumors; READ, 3 controls vs. 161 tumors) and mRNA (COAD, 41 controls vs. 478 tumors; READ, 10 controls vs. 166 tumors) data from The Cancer Genome Atlas (TCGA) database and then used DESeq2, RegParallel, miRDB, TargetScanHuman 7.2, DAVID 6.8, STRING, and Cytoscape software to identify the potential prognosis biomarkers.
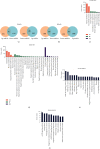
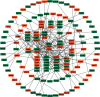
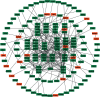
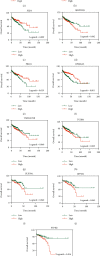
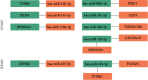
Results: We identified 175 differential expression miRNAs (DEMs) and 3747 differential expression genes (DEGs) in COAD and 184 DEMs and 3928 DEGs in READ. And then, we obtained 21 (13 in COAD and 8 in READ) DEMs associated with the survival rates, which correlated with 440 (217 in COAD and 223 in READ) overlapping DEGs. Through survival analysis for those overlapping DEGs, we found 11 (8 in COAD and 3 in READ) overlapping DGEs associated with survival rates of patients, which were correlated with 9 (7 in COAD and 2 in READ) DEMs significantly.
Conclusion: In this study, we found several candidate prognostic biomarkers which have been identified in various cancers and also found several new prognosis biomarkers of COAD and READ. In conclusion, this analysis based on theoretical knowledge and clinical outcomes we have done needs further confirmation by more researches.
Copyright © 2020 Xiao-Liang Xing et al.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Colon Cancer Laparoscopic or Open Resection Study Group, Buunen M., Veldkamp R., et al. Survival after laparoscopic surgery versus open surgery for colon cancer: long-term outcome of a randomised clinical trial. The Lancet Oncology. 2009;10(1):44–52. doi: 10.1016/S1470-2045(08)70310-3. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

