Altered Capicua expression drives regional Purkinje neuron vulnerability through ion channel gene dysregulation in spinocerebellar ataxia type 1
- PMID: 32964235
- PMCID: PMC7689299
- DOI: 10.1093/hmg/ddaa212
Altered Capicua expression drives regional Purkinje neuron vulnerability through ion channel gene dysregulation in spinocerebellar ataxia type 1
Abstract
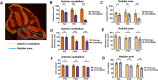
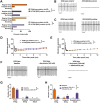
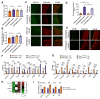
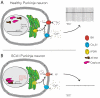
Selective neuronal vulnerability in neurodegenerative disease is poorly understood. Using the ATXN1[82Q] model of spinocerebellar ataxia type 1 (SCA1), we explored the hypothesis that regional differences in Purkinje neuron degeneration could provide novel insights into selective vulnerability. ATXN1[82Q] Purkinje neurons from the anterior cerebellum were found to degenerate earlier than those from the nodular zone, and this early degeneration was associated with selective dysregulation of ion channel transcripts and altered Purkinje neuron spiking. Efforts to understand the basis for selective dysregulation of channel transcripts revealed modestly increased expression of the ATXN1 co-repressor Capicua (Cic) in anterior cerebellar Purkinje neurons. Importantly, disrupting the association between ATXN1 and Cic rescued the levels of these ion channel transcripts, and lentiviral overexpression of Cic in the nodular zone accelerated both aberrant Purkinje neuron spiking and neurodegeneration. These findings reinforce the central role for Cic in SCA1 cerebellar pathophysiology and suggest that only modest reductions in Cic are needed to have profound therapeutic impact in SCA1.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


Similar articles
-
ATXN1-CIC Complex Is the Primary Driver of Cerebellar Pathology in Spinocerebellar Ataxia Type 1 through a Gain-of-Function Mechanism.Neuron. 2018 Mar 21;97(6):1235-1243.e5. doi: 10.1016/j.neuron.2018.02.013. Epub 2018 Mar 8. Neuron. 2018. PMID: 29526553 Free PMC article.
-
Sphingolipid metabolism governs Purkinje cell patterned degeneration in Atxn1[82Q]/+ mice.Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2016969118. doi: 10.1073/pnas.2016969118. Proc Natl Acad Sci U S A. 2021. PMID: 34479994 Free PMC article.
-
Increased intrinsic membrane excitability is associated with olivary hypertrophy in spinocerebellar ataxia type 1.Hum Mol Genet. 2024 Dec 6;33(24):2159-2176. doi: 10.1093/hmg/ddae146. Hum Mol Genet. 2024. PMID: 39475127 Free PMC article.
-
Therapeutic Strategies for Spinocerebellar Ataxia Type 1.Biomolecules. 2023 May 2;13(5):788. doi: 10.3390/biom13050788. Biomolecules. 2023. PMID: 37238658 Free PMC article. Review.
-
Cholecystokinin Activation of Cholecystokinin 1 Receptors: a Purkinje Cell Neuroprotective Pathway.Cerebellum. 2023 Aug;22(4):756-760. doi: 10.1007/s12311-022-01428-x. Epub 2022 Jun 23. Cerebellum. 2023. PMID: 35733029 Free PMC article. Review.
Cited by
-
Single nuclei RNA sequencing investigation of the Purkinje cell and glial changes in the cerebellum of transgenic Spinocerebellar ataxia type 1 mice.Front Cell Neurosci. 2022 Nov 15;16:998408. doi: 10.3389/fncel.2022.998408. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36457352 Free PMC article.
-
Discovery of Novel Activators of Large-Conductance Calcium-Activated Potassium Channels for the Treatment of Cerebellar Ataxia.Mol Pharmacol. 2022 Jul;102(1):438-449. doi: 10.1124/molpharm.121.000478. Epub 2022 Apr 30. Mol Pharmacol. 2022. PMID: 35489717 Free PMC article.
-
Elevation of hsa-miR-7-5p level mediated by CtBP1-p300-AP1 complex targets ATXN1 to trigger NF-κB-dependent inflammation response.J Mol Med (Berl). 2023 Mar;101(3):223-235. doi: 10.1007/s00109-022-02274-4. Epub 2023 Jan 11. J Mol Med (Berl). 2023. PMID: 36629882
-
Cerebellar Regional Dissection for Molecular Analysis.J Vis Exp. 2020 Dec 5;(166):10.3791/61922. doi: 10.3791/61922. J Vis Exp. 2020. PMID: 33346205 Free PMC article.
-
Loss of Flocculus Purkinje Cell Firing Precision Leads to Impaired Gaze Stabilization in a Mouse Model of Spinocerebellar Ataxia Type 6 (SCA6).Cells. 2022 Sep 2;11(17):2739. doi: 10.3390/cells11172739. Cells. 2022. PMID: 36078147 Free PMC article.
References
-
- Saxena S. and Caroni P. (2011) Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron, 71, 35–48. - PubMed
-
- Orr H.T., Chung M.Y., Banfi S., Kwiatkowski T.J. Jr., Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P. and Zoghbi H.Y. (1993) Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet., 4, 221–226. - PubMed
-
- Rub U., Burk K., Timmann D., Dunnen W., Seidel K., Farrag K., Brunt E., Heinsen H., Egensperger R., Bornemann A. et al. (2012) Spinocerebellar ataxia type 1 (SCA1): new pathoanatomical and clinico-pathological insights. Neuropathol. Appl. Neurobiol., 38, 665–680. - PubMed
-
- Servadio A., Koshy B., Armstrong D., Antalffy B., Orr H.T. and Zoghbi H.Y. (1995) Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nat. Genet., 10, 94–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

