Identification of destabilizing SNPs in SARS-CoV2-ACE2 protein and spike glycoprotein: implications for virus entry mechanisms
- PMID: 32964802
- PMCID: PMC7544926
- DOI: 10.1080/07391102.2020.1823885
Identification of destabilizing SNPs in SARS-CoV2-ACE2 protein and spike glycoprotein: implications for virus entry mechanisms
Abstract
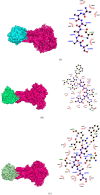
COVID-19 an outbreak of a novel corona virus originating from Wuhan, China in December 2019 has now spread across the entire world and has been declared a pandemic by WHO. Angiotensin converting enzyme 2 (ACE2) is a receptor protein that interacts with the spike glycoprotein of the host to facilitate the entry of coronavirus (SARS-CoV-2) hence causing the disease (COVID-19). Our experimental design is based on bioinformatics approach that combines sequence, structure and consensus based tools to label a protein coding single nucleotide polymorphism (SNP) as damaging/deleterious or neutral. The interaction of wildtype ACE2-spike glycoprotein and their variants were analyzed using docking studies. The mutations W461R, G405E and F588S in ACE2 receptor protein and population specific mutations P391S, C12S and G1223A in the spike glycoprotein were predicted as highly destabilizing to the structure of the bound complex. So far, no extensive in silico study has been reported that identifies the effect of SNPs on Spike glycoprotein-ACE2 interaction exploring both sequence and structural features. To this end, this study conducted an in-depth analysis that facilitates in identifying the mutations that blocks the interaction of two proteins that can result in stopping the virus from entering the host cell.Communicated by Ramaswamy H. Sarma.
Keywords: 2019-nCOV; ACE2; COVID-19; SNPs; spike glycoprotein.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
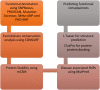


References
-
- Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., Martin, M. J., Natale, D. A., O'Donovan, C., Redaschi, N., & Yeh, L.-S L. (2004). UniProt: The universal protein knowledgebase. Nucleic Acids Research, 32(Database issue), D115–9. 10.1093/nar/gkh131 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
