Transposon clusters as substrates for aberrant splice-site activation
- PMID: 32965162
- PMCID: PMC7951965
- DOI: 10.1080/15476286.2020.1805909
Transposon clusters as substrates for aberrant splice-site activation
Abstract
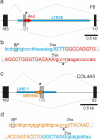
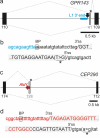
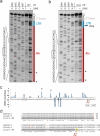
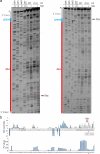
Transposed elements (TEs) have dramatically shaped evolution of the exon-intron structure and significantly contributed to morbidity, but how recent TE invasions into older TEs cooperate in generating new coding sequences is poorly understood. Employing an updated repository of new exon-intron boundaries induced by pathogenic mutations, termed DBASS, here we identify novel TE clusters that facilitated exon selection. To explore the extent to which such TE exons maintain RNA secondary structure of their progenitors, we carried out structural studies with a composite exon that was derived from a long terminal repeat (LTR78) and AluJ and was activated by a C > T mutation optimizing the 5' splice site. Using a combination of SHAPE, DMS and enzymatic probing, we show that the disease-causing mutation disrupted a conserved AluJ stem that evolved from helix 3.3 (or 5b) of 7SL RNA, liberating a primordial GC 5' splice site from the paired conformation for interactions with the spliceosome. The mutation also reduced flexibility of conserved residues in adjacent exon-derived loops of the central Alu hairpin, revealing a cross-talk between traditional and auxilliary splicing motifs that evolved from opposite termini of 7SL RNA and were approximated by Watson-Crick base-pairing already in organisms without spliceosomal introns. We also identify existing Alu exons activated by the same RNA rearrangement. Collectively, these results provide valuable TE exon models for studying formation and kinetics of pre-mRNA building blocks required for splice-site selection and will be useful for fine-tuning auxilliary splicing motifs and exon and intron size constraints that govern aberrant splice-site activation.
Keywords: DBASS3; DBASS5; RNA processing; RNA secondary structure; Transposed element; genetic disease; lariat branch point; mutation; splice site.
Conflict of interest statement
None. Inventor royalties from a licensing agreement for the intellectual property unrelated to this work (US patents 9,714,422 and 10,196,639) were personally contributed to the University of Southampton Faculty of Medicine and converted into a research grant administered by the same institution.
Figures


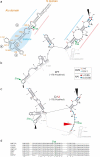
Similar articles
-
DBASS3 and DBASS5: databases of aberrant 3'- and 5'-splice sites.Nucleic Acids Res. 2011 Jan;39(Database issue):D86-91. doi: 10.1093/nar/gkq887. Epub 2010 Oct 6. Nucleic Acids Res. 2011. PMID: 20929868 Free PMC article.
-
Transposable elements in disease-associated cryptic exons.Hum Genet. 2010 Feb;127(2):135-54. doi: 10.1007/s00439-009-0752-4. Epub 2009 Oct 10. Hum Genet. 2010. PMID: 19823873
-
The role of short RNA loops in recognition of a single-hairpin exon derived from a mammalian-wide interspersed repeat.RNA Biol. 2015;12(1):54-69. doi: 10.1080/15476286.2015.1017207. RNA Biol. 2015. PMID: 25826413 Free PMC article.
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
-
Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum.Mutat Res. 1998 Nov;411(3):179-214. doi: 10.1016/s1383-5742(98)00013-1. Mutat Res. 1998. PMID: 9804951 Review.
Cited by
-
Alu RNA fold links splicing with signal recognition particle proteins.Nucleic Acids Res. 2023 Aug 25;51(15):8199-8216. doi: 10.1093/nar/gkad500. Nucleic Acids Res. 2023. PMID: 37309897 Free PMC article.
-
Background splicing as a predictor of aberrant splicing in genetic disease.RNA Biol. 2022;19(1):256-265. doi: 10.1080/15476286.2021.2024031. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35188075 Free PMC article.
-
Dynamic Variations of 3'UTR Length Reprogram the mRNA Regulatory Landscape.Biomedicines. 2021 Oct 28;9(11):1560. doi: 10.3390/biomedicines9111560. Biomedicines. 2021. PMID: 34829789 Free PMC article. Review.
-
Characterization of SOD1-DT, a Divergent Long Non-Coding RNA in the Locus of the SOD1 Human Gene.Cells. 2023 Aug 13;12(16):2058. doi: 10.3390/cells12162058. Cells. 2023. PMID: 37626868 Free PMC article.
-
Introns: the "dark matter" of the eukaryotic genome.Front Genet. 2023 May 16;14:1150212. doi: 10.3389/fgene.2023.1150212. eCollection 2023. Front Genet. 2023. PMID: 37260773 Free PMC article. Review.
References
-
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. - PubMed
-
- Wahl MC, Will CL, Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Wang Z, Rolish ME, Yeo G, et al. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
