Targeting Polyamines Inhibits Coronavirus Infection by Reducing Cellular Attachment and Entry
- PMID: 32966040
- PMCID: PMC7539557
- DOI: 10.1021/acsinfecdis.0c00491
Targeting Polyamines Inhibits Coronavirus Infection by Reducing Cellular Attachment and Entry
Abstract
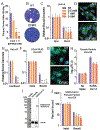
Coronaviruses first garnered widespread attention in 2002 when the severe acute respiratory syndrome coronavirus (SARS-CoV) emerged from bats in China and rapidly spread in human populations. Since then, Middle East respiratory syndrome coronavirus (MERS-CoV) emerged and still actively infects humans. The recent SARS-CoV-2 outbreak and the resulting disease (coronavirus disease 2019, COVID19) have rapidly and catastrophically spread and highlighted significant limitations to our ability to control and treat infection. Thus, a basic understanding of entry and replication mechanisms of coronaviruses is necessary to rationally evaluate potential antivirals. Here, we show that polyamines, small metabolites synthesized in human cells, facilitate coronavirus replication and the depletion of polyamines with FDA-approved molecules significantly reduces coronavirus replication. We find that diverse coronaviruses, including endemic and epidemic coronaviruses, exhibit reduced attachment and entry into polyamine-depleted cells. We further demonstrate that several molecules targeting the polyamine biosynthetic pathway are antiviral in vitro. In sum, our data suggest that polyamines are critical to coronavirus replication and represent a highly promising drug target in the current and any future coronavirus outbreaks.
Keywords: SARS-CoV-2; binding; coronavirus; polyamines.
Figures

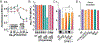
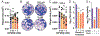

References
-
- WHO | Middle East respiratory syndrome coronavirus (MERS-CoV) http://www.who.int/emergencies/mers-cov/en/ (accessed Jun 6, 2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

