Replication Stress Induces Global Chromosome Breakage in the Fragile X Genome
- PMID: 32966779
- PMCID: PMC7549430
- DOI: 10.1016/j.celrep.2020.108179
Replication Stress Induces Global Chromosome Breakage in the Fragile X Genome
Erratum in
-
Replication stress induces global chromosome breakage in the fragile X genome.Cell Rep. 2021 Mar 23;34(12):108838. doi: 10.1016/j.celrep.2021.108838. Cell Rep. 2021. PMID: 33761342 Free PMC article. No abstract available.
Abstract
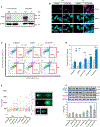
Fragile X syndrome (FXS) is a neurodevelopmental disorder caused by mutations in the FMR1 gene and deficiency of a functional FMRP protein. FMRP is known as a translation repressor whose nuclear function is not understood. We investigated the global impact on genome stability due to FMRP loss. Using Break-seq, we map spontaneous and replication stress-induced DNA double-strand breaks (DSBs) in an FXS patient-derived cell line. We report that the genomes of FXS cells are inherently unstable and accumulate twice as many DSBs as those from an unaffected control. We demonstrate that replication stress-induced DSBs in FXS cells colocalize with R-loop forming sequences. Exogenously expressed FMRP in FXS fibroblasts ameliorates DSB formation. FMRP, not the I304N mutant, abates R-loop-induced DSBs during programmed replication-transcription conflict. These results suggest that FMRP is a genome maintenance protein that prevents R-loop accumulation. Our study provides insights into the etiological basis for FXS.
Keywords: DNA double-strand breaks; DNA replication stress; DSB; FMRP; FXS; I304N; R-loops; chromosome fragile sites; fragile X syndrome; genome instability.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

