Chromatin Landscape Underpinning Human Dendritic Cell Heterogeneity
- PMID: 32966789
- PMCID: PMC7546547
- DOI: 10.1016/j.celrep.2020.108180
Chromatin Landscape Underpinning Human Dendritic Cell Heterogeneity
Abstract
Human dendritic cells (DCs) comprise subsets with distinct phenotypic and functional characteristics, but the transcriptional programs that dictate their identity remain elusive. Here, we analyze global chromatin accessibility profiles across resting and stimulated human DC subsets by means of the assay for transposase-accessible chromatin using sequencing (ATAC-seq). We uncover specific regions of chromatin accessibility for each subset and transcriptional regulators of DC function. By comparing plasmacytoid DC responses to IFN-I-producing and non-IFN-I-producing conditions, we identify genetic programs related to their function. Finally, by intersecting chromatin accessibility with genome-wide association studies, we recognize DC subset-specific enrichment of heritability in autoimmune diseases. Our results unravel the basis of human DC subset heterogeneity and provide a framework for their analysis in disease pathogenesis.
Keywords: ASDCs; ATAC-seq; AXL+ DCs; IFN-I; cDC1; cDC2; dendritic cells; human; monocytes; pDCs; plasmacytoid dendritic cells; tDCs; transitional dendritic cells.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests H.Y.C. is affiliated with Accent Therapeutics, Boundless Bio, 10x Genomics, Arsenal Biosciences, and Spring Discovery. A.T.S. is a scientific co-founder of Immunai and receives research funding from Arsenal Biosciences.
Figures






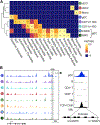
References
-
- Alculumbre SG, Saint-André V, Di Domizio J, Vargas P, Sirven P, Bost P, Maurin M, Maiuri P, Wery M, Roman MS, et al. (2018). Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol 19, 63–75. - PubMed
-
- Anderson KL, Perkin H, Surh CD, Venturini S, Maki RA, and Torbett BE (2000). Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J. Immunol 164, 1855–1861. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

