A Membrane-Tethered Ubiquitination Pathway Regulates Hedgehog Signaling and Heart Development
- PMID: 32966817
- PMCID: PMC7686252
- DOI: 10.1016/j.devcel.2020.08.012
A Membrane-Tethered Ubiquitination Pathway Regulates Hedgehog Signaling and Heart Development
Abstract
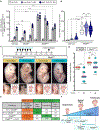
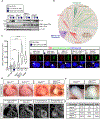
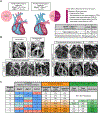
The etiology of congenital heart defects (CHDs), which are among the most common human birth defects, is poorly understood because of its complex genetic architecture. Here, we show that two genes implicated in CHDs, Megf8 and Mgrn1, interact genetically and biochemically to regulate the strength of Hedgehog signaling in target cells. MEGF8, a transmembrane protein, and MGRN1, a RING superfamily E3 ligase, assemble to form a receptor-like ubiquitin ligase complex that catalyzes the ubiquitination and degradation of the Hedgehog pathway transducer Smoothened. Homozygous Megf8 and Mgrn1 mutations increased Smoothened abundance and elevated sensitivity to Hedgehog ligands. While mice heterozygous for loss-of-function Megf8 or Mgrn1 mutations were normal, double heterozygous embryos exhibited an incompletely penetrant syndrome of CHDs with heterotaxy. Thus, genetic interactions can arise from biochemical mechanisms that calibrate morphogen signaling strength, a conclusion broadly relevant for the many human diseases in which oligogenic inheritance is emerging as a mechanism for heritability.
Keywords: Hedgehog signaling; Smoothened; congenital heart disease; heart development; heterotaxy; left-right patterning; morphogen; oligogenic inheritance; primary cilia; ubiquitin.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




Comment in
-
Ubiquitin Tunes Hedgehog in Matters of the Heart.Dev Cell. 2020 Nov 23;55(4):385-386. doi: 10.1016/j.devcel.2020.10.017. Dev Cell. 2020. PMID: 33232673
References
-
- Aune CN, Chatterjee B, Zhao X-Q, Francis R, Bracero L, Yu Q, Rosenthal J, Leatherbury L, and Lo CW (2008). Mouse model of heterotaxy with single ventricle spectrum of cardiac anomalies. Pediatr. Res 63, 9–14. - PubMed
-
- Badano JL, Mitsuma N, Beales PL, and Katsanis N (2006). The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet 7, 125–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

