Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production-Reproductive Traits
- PMID: 32967074
- PMCID: PMC7552765
- DOI: 10.3390/ani10091704
Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production-Reproductive Traits
Abstract
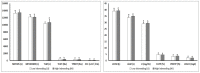
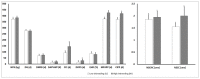
The main objective of this study was to analyze the effects of the inbreeding degree in high-producing primiparous dairy cows genotypically and phenotypically evaluated and its impacts on production and reproductive parameters. Eighty Holstein-Friesian primiparous cows (age: ~26 months; ~450 kg body weight) were previously genomically analyzed to determine the Inbreeding Index (II) and were divided into two groups: low inbreeding group (LI: <2.5; n = 40) and high inbreeding group (HI: ≥2.5 and ≤5.0; n = 40). Genomic determinations of production and reproductive parameters (14 in total), together with analyses of production (12) and reproductive (11) phenotypic parameters (23 in total) were carried out. Statistically significant differences were obtained between groups concerning the genomic parameters of Milk Production at 305 d and Protein Production at 305 d and the reproductive parameter Daughter Calving Ease, the first two being higher in cows of the HI group and the third lower in the LI group (p < 0.05). For the production phenotypic parameters, statistically significant differences were observed between both groups in the Total Fat, Total Protein, and Urea parameters, the first two being higher in the LI group (p < 0.05). Also, significant differences were observed in several reproductive phenotypic parameters, such as Number of Services per Conception, Calving to Conception Interval, Days Open Post Service, and Current Inter-Partum Period, all of which negatively influenced the HI group (p < 0.05). In addition, correlation analyses were performed between production and reproductive genomic parameters separately and in each consanguinity group. The results showed multiple positive and negative correlations between the production and reproductive parameters independently of the group analyzed, being these correlations more remarkable for the reproductive parameters in the LI group and the production parameters in the HI group (p < 0.05). In conclusion, the degree of inbreeding significantly influenced the results, affecting different genomic and phenotypic production and reproductive parameters in high-producing primiparous cows. The determination of the II in first-calf heifers is crucial to evaluate the negative effects associated with homozygosity avoiding an increase in inbreeding depression on production and reproductive traits.
Keywords: dairy cattle; genomic analysis; genotypic parameters; homozygosis; inbreeding index; phenotypic traits; primiparous; production; reproductive performance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- García-Ruiz A., Cole J.B., VanRaden P.M., Wiggans G.R., Ruiz-López F.J., Van Tassell C.P. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. USA. 2016;113:E3995–E4004. doi: 10.1073/pnas.1519061113. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

