G-Quadruplexes in the Archaea Domain
- PMID: 32967357
- PMCID: PMC7565180
- DOI: 10.3390/biom10091349
G-Quadruplexes in the Archaea Domain
Abstract
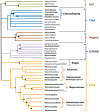
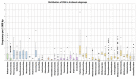
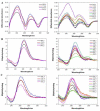
The importance of unusual DNA structures in the regulation of basic cellular processes is an emerging field of research. Amongst local non-B DNA structures, G-quadruplexes (G4s) have gained in popularity during the last decade, and their presence and functional relevance at the DNA and RNA level has been demonstrated in a number of viral, bacterial, and eukaryotic genomes, including humans. Here, we performed the first systematic search of G4-forming sequences in all archaeal genomes available in the NCBI database. In this article, we investigate the presence and locations of G-quadruplex forming sequences using the G4Hunter algorithm. G-quadruplex-prone sequences were identified in all archaeal species, with highly significant differences in frequency, from 0.037 to 15.31 potential quadruplex sequences per kb. While G4 forming sequences were extremely abundant in Hadesarchaea archeon (strikingly, more than 50% of the Hadesarchaea archaeon isolate WYZ-LMO6 genome is a potential part of a G4-motif), they were very rare in the Parvarchaeota phylum. The presence of G-quadruplex forming sequences does not follow a random distribution with an over-representation in non-coding RNA, suggesting possible roles for ncRNA regulation. These data illustrate the unique and non-random localization of G-quadruplexes in Archaea.
Keywords: Archaea; G4-forming motif; genome analysis; sequence prediction; unusual nucleic acid structures.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

