Comparative transcriptome analysis revealed the cooperative regulation of sucrose and IAA on adventitious root formation in lotus (Nelumbo nucifera Gaertn)
- PMID: 32967611
- PMCID: PMC7510093
- DOI: 10.1186/s12864-020-07046-3
Comparative transcriptome analysis revealed the cooperative regulation of sucrose and IAA on adventitious root formation in lotus (Nelumbo nucifera Gaertn)
Abstract
Background: In China, lotus is an important cultivated crop with multiple applications in ornaments, food, and environmental purification. Adventitious roots (ARs), a secondary root is necessary for the uptake of nutrition and water as the lotus principle root is underdeveloped. Therefore, AR formation in seedlings is very important for lotus breeding due to its effect on plant early growth. As lotus ARs formation was significantly affected by sucrose treatment, we analyzed the expression of genes and miRNAs upon treatment with differential concentrations of sucrose, and a crosstalk between sucrose and IAA was also identified.
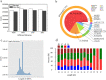
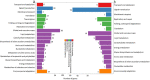
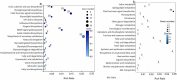
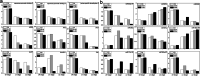
Results: Notably, 20 mg/L sucrose promoted the ARs development, whereas 60 mg/L sucrose inhibited the formation of ARs. To investigate the regulatory pathway during ARs formation, the expression of genes and miRNAs was evaluated by high-throughput tag-sequencing. We observed that the expression of 5438, 5184, and 5345 genes was enhanced in the GL20/CK0, GL60/CK0, and CK1/CK0 libraries, respectively. Further, the expression of 73, 78, and 71 miRNAs was upregulated in the ZT20/MCK0, ZT60/MCK0, and MCK1/MCK0 libraries, respectively. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that most of the differentially expressed genes and miRNAs in the GL20/GL60 and ZT20/ZT60 libraries were involved in signal transduction. A large number of these genes (29) and miRNAs (53) were associated with plant hormone metabolism. We observed an association between five miRNAs (miR160, miR156a-5p, miR397-5p_1, miR396a and miR167d) and nine genes (auxin response factor, protein brassinosteroid insensitive 1, laccase, and peroxidase 27) in the ZT20/ ZT60 libraries during ARs formation. Quantitative polymerase chain reaction (qRT-PCR) was used to validate the high-throughput tag-sequencing data.
Conclusions: We found that the expression of many critical genes involved in IAA synthesis and IAA transport was changed after treatment with various concentration of sucrose. Based on the change of these genes expression, IAA and sucrose content, we concluded that sucrose and IAA cooperatively regulated ARs formation. Sucrose affected ARs formation by improving IAA content at induction stage, and increased sucrose content might be also required for ARs development according to the changes tendency after application of exogenous IAA.
Keywords: Adventitious roots; Gene; Lotus; Sucrose; miRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Shen-Miller J. Sacred lotus, the long-living fruits of China antique. Seed Sci Res. 2002;12:131–143. doi: 10.1079/SSR2002112. - DOI
-
- Cheng LB, Liu HY, Jiang RZ, Li SY. A proteomics analysis of adventitious root formation after leaf removal in lotus (Nelumbo nucifera Gaertn.) ZNC. 2018;73:375–389. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

