B chromosomes of multiple species have intense evolutionary dynamics and accumulated genes related to important biological processes
- PMID: 32967626
- PMCID: PMC7509943
- DOI: 10.1186/s12864-020-07072-1
B chromosomes of multiple species have intense evolutionary dynamics and accumulated genes related to important biological processes
Abstract
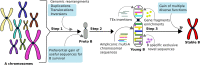
Background: One of the biggest challenges in chromosome biology is to understand the occurrence and complex genetics of the extra, non-essential karyotype elements, commonly known as supernumerary or B chromosomes (Bs). The non-Mendelian inheritance and non-pairing abilities of B chromosomes make them an interesting model for genomics studies, thus bringing to bear different questions about their genetic composition, evolutionary survival, maintenance and functional role inside the cell. This study uncovers these phenomena in multiple species that we considered as representative organisms of both vertebrate and invertebrate models for B chromosome analysis.
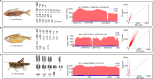
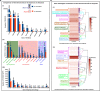
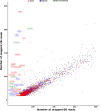
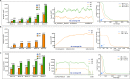
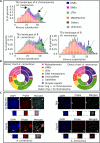
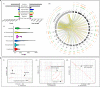
Results: We sequenced the genomes of three animal species including two fishes Astyanax mexicanus and Astyanax correntinus, and a grasshopper Abracris flavolineata, each with and without Bs, and identified their B-localized genes and repeat contents. We detected unique sequences occurring exclusively on Bs and discovered various evolutionary patterns of genomic rearrangements associated to Bs. In situ hybridization and quantitative polymerase chain reactions further validated our genomic approach confirming detection of sequences on Bs. The functional annotation of B sequences showed that the B chromosome comprises regions of gene fragments, novel genes, and intact genes, which encode a diverse set of functions related to important biological processes such as metabolism, morphogenesis, reproduction, transposition, recombination, cell cycle and chromosomes functions which might be important for their evolutionary success.
Conclusions: This study reveals the genomic structure, composition and function of Bs, which provide new insights for theories of B chromosome evolution. The selfish behavior of Bs seems to be favored by gained genes/sequences.
Keywords: Evolution; Extra chromosome; Genome; Next generation sequencing; Supernumerary chromosome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Comment in
-
Irreproducible results and unsupported conclusions in Ahmad et al. [BMC genomics (2020) 21:656].BMC Genomics. 2023 Dec 18;24(1):778. doi: 10.1186/s12864-023-09883-4. BMC Genomics. 2023. PMID: 38105238 Free PMC article. No abstract available.
References
-
- Longley AE. Supernumerary chromosomes in Zea mays. J Agric Res. 1927;35:769–784.
-
- Wilson EB. Studies on chromosomes. V. the chromosomes of metapodius. A contribution to the hypothesis of the genetic continuity of chromosomes. J Exp Zool. 1909;6:147–205. doi: 10.1002/jez.1400060202. - DOI
-
- Camacho JPM. B chromosomes. In: Gregory TR, editor. The evolution of the genome. 2005. pp. 223–286.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

