A Novel Platform Using RNA Signatures To Accelerate Antimicrobial Susceptibility Testing in Neisseria gonorrhoeae
- PMID: 32967905
- PMCID: PMC7685871
- DOI: 10.1128/JCM.01152-20
A Novel Platform Using RNA Signatures To Accelerate Antimicrobial Susceptibility Testing in Neisseria gonorrhoeae
Erratum in
-
Erratum for Hashemi et al., "A Novel Platform Using RNA Signatures To Accelerate Antimicrobial Susceptibility Testing in Neisseria gonorrhoeae".J Clin Microbiol. 2021 Feb 18;59(3):e03041-20. doi: 10.1128/JCM.03041-20. Print 2021 Feb 18. J Clin Microbiol. 2021. PMID: 33826529 Free PMC article. No abstract available.
Abstract
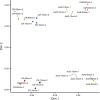
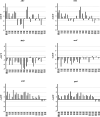
The rise of antimicrobial-resistant pathogens can be attributed to the lack of a rapid pathogen identification (ID) or antimicrobial susceptibility testing (AST), resulting in delayed therapeutic decisions at the point of care. Gonorrhea is usually empirically treated, with no AST results available before treatment, thus contributing to the rapid rise in drug resistance. Here, we present a rapid AST platform using RNA signatures for Neisseria gonorrhoeae Transcriptome sequencing (RNA-seq) followed by bioinformatic tools was applied to explore potential markers in the transcriptome profile of N. gonorrhoeae upon minutes of azithromycin exposure. Validation of candidate markers using quantitative real-time PCR (qRT-PCR) showed that two markers (arsR [NGO1562] and rpsO) can deliver accurate AST results across 14 tested isolates. Further validation of our susceptibility threshold in comparison to MIC across 64 more isolates confirmed the reliability of our platform. Our RNA markers combined with emerging molecular point-of-care systems has the potential to greatly accelerate both ID and AST to inform treatment.
Keywords: Neisseria gonorrhoeae; RNA markers; RNA-seq; antimicrobial susceptibility testing; azithromycin.
Copyright © 2020 American Society for Microbiology.
Figures




References
-
- Centers for Disease Control and Prevention. 2000. Gonorrhea—United States, 1998. MMWR Morb Mortal Wkly Rep 49:538–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

