Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR
- PMID: 32968075
- PMCID: PMC7511968
- DOI: 10.1038/s41467-020-18611-5
Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR
Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is commonly diagnosed by reverse transcription polymerase chain reaction (RT-PCR) to detect viral RNA in patient samples, but RNA extraction constitutes a major bottleneck in current testing. Methodological simplification could increase diagnostic availability and efficiency, benefitting patient care and infection control. Here, we describe methods circumventing RNA extraction in COVID-19 testing by performing RT-PCR directly on heat-inactivated or lysed samples. Our data, including benchmarking using 597 clinical patient samples and a standardised diagnostic system, demonstrate that direct RT-PCR is viable option to extraction-based tests. Using controlled amounts of active SARS-CoV-2, we confirm effectiveness of heat inactivation by plaque assay and evaluate various generic buffers as transport medium for direct RT-PCR. Significant savings in time and cost are achieved through RNA-extraction-free protocols that are directly compatible with established PCR-based testing pipelines. This could aid expansion of COVID-19 testing.
Conflict of interest statement
The authors declare no competing interests.
Figures



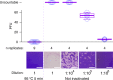

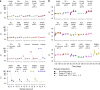

References
-
- Taipale, J., Romer, P. & Linnarsson, S. Population-scale testing can suppress the spread of COVID-19. medRxiv.org, 2020.04.27.20078329 (2020).
-
- Ferguson, N. M. et al. Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. Imperial College COVID-19 Response Team; https://www.imperial.ac.uk/media/imperial-college/medicine/sph/ide/gida-... (2020). - PMC - PubMed
-
- Corman VM, et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance. 2012;17:2–10. - PubMed
-
- Centers for Disease Control and Prevention. Real-Time RT-PCR Panel for Detection 2019-Novel Coronavirus. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-for-det... (2020).
Publication types
MeSH terms
Substances
Grants and funding
- 2017-01723/Vetenskapsrådet (Swedish Research Council)/International
- M16/17/Ragnar Söderbergs stiftelse (Ragnar Söderberg Foundation)/International
- 2020.0182/Science for Life Laboratory (SciLifeLab)/International
- 2020.0182/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

