Complex genetic encoding of the hepatitis B virus on-drug persistence
- PMID: 32968103
- PMCID: PMC7511938
- DOI: 10.1038/s41598-020-72467-9
Complex genetic encoding of the hepatitis B virus on-drug persistence
Abstract
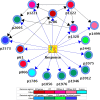
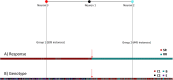
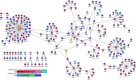
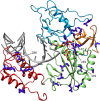
Tenofovir disoproxil fumarate (TDF) is one of the nucleotide analogs capable of inhibiting the reverse transcriptase (RT) activity of HIV and hepatitis B virus (HBV). There is no known HBV resistance to TDF. However, detectable variation in duration of HBV persistence in patients on TDF therapy suggests the existence of genetic mechanisms of on-drug persistence that reduce TDF efficacy for some HBV strains without affording actual resistance. Here, the whole genome of intra-host HBV variants (N = 1,288) was sequenced from patients with rapid (RR, N = 5) and slow response (SR, N = 5) to TDF. Association of HBV genomic and protein polymorphic sites to RR and SR was assessed using phylogenetic analysis and Bayesian network methods. We show that, in difference to resistance to nucleotide analogs, which is mainly associated with few specific mutations in RT, the HBV on-TDF persistence is defined by genetic variations across the entire HBV genome. Analysis of the inferred 3D-structures indicates no difference in affinity of TDF binding by RT encoded by intra-host HBV variants that rapidly decline or persist in presence of TDF. This finding suggests that effectiveness of TDF recognition and binding does not contribute significantly to on-drug persistence. Differences in patterns of genetic associations to TDF response between HBV genotypes B and C and lack of a single pattern of mutations among intra-host variants sensitive to TDF indicate a complex genetic encoding of the trait. We hypothesize that there are many genetic mechanisms of on-drug persistence, which are differentially available to HBV strains. These pervasive mechanisms are insufficient to prevent viral inhibition completely but may contribute significantly to robustness of actual resistance. On-drug persistence may reduce the overall effectiveness of therapy and should be considered for development of more potent drugs.
Conflict of interest statement
H.T., J.L., X.X., G.X., L.G.-R. and Y.K. (Centers for Disease Control and Prevention) declare no competing interests. CDC Disclaimer: the findings and conclusions of this manuscript are those of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention. K.K. was an employee and stockholder at Gilead Sciences at the time the work was completed. A.G. is an employee and stockholder at Gilead Sciences. H.L.Y.C. is an advisor of Abbvie, Aligos, Arbutus, Gilead Sciences, Intellia, Janssen, MedImmune, Merck, Roche, Vir Biotechnology, Vaccitech, VenatoRx; and a speaker for Gilead Sciences, Mylan and Roche.
Figures

Similar articles
-
Variability in the Responses of Hepatitis B Virus D-Subgenotypes to Antiviral Therapy: Designing Pan-D-Subgenotypic Reverse Transcriptase Inhibitors.J Virol. 2022 Jan 26;96(2):e0180021. doi: 10.1128/JVI.01800-21. Epub 2021 Nov 3. J Virol. 2022. PMID: 34730399 Free PMC article.
-
In vitro susceptibility of lamivudine-resistant hepatitis B virus to adefovir and tenofovir.Antivir Ther. 2004 Jun;9(3):353-63. Antivir Ther. 2004. PMID: 15259898
-
Prolonged use of tenofovir in HIV/hepatitis B virus (HBV)-coinfected individuals does not lead to HBV polymerase mutations and is associated with persistence of lamivudine HBV polymerase mutations.HIV Med. 2009 Apr;10(4):229-35. doi: 10.1111/j.1468-1293.2008.00675.x. Epub 2009 Jan 28. HIV Med. 2009. PMID: 19178592
-
Development of tenofovir disoproxil fumarate resistance after complete viral suppression in a patient with treatment-naïve chronic hepatitis B: A case report and review of the literature.World J Gastroenterol. 2018 May 7;24(17):1919-1924. doi: 10.3748/wjg.v24.i17.1919. World J Gastroenterol. 2018. PMID: 29740207 Free PMC article. Review.
-
Naturally occurring hepatitis B virus reverse transcriptase mutations related to potential antiviral drug resistance and liver disease progression.World J Gastroenterol. 2018 Apr 28;24(16):1708-1724. doi: 10.3748/wjg.v24.i16.1708. World J Gastroenterol. 2018. PMID: 29713126 Free PMC article. Review.
Cited by
-
Molecular Mechanisms during Hepatitis B Infection and the Effects of the Virus Variability.Viruses. 2021 Jun 18;13(6):1167. doi: 10.3390/v13061167. Viruses. 2021. PMID: 34207116 Free PMC article. Review.
-
Should We Treat Immune Tolerant Chronic Hepatitis B? Lessons from Asia.J Clin Exp Hepatol. 2022 Jan-Feb;12(1):144-154. doi: 10.1016/j.jceh.2021.08.023. Epub 2021 Aug 27. J Clin Exp Hepatol. 2022. PMID: 35068795 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

