Bardoxolone conjugation enables targeted protein degradation of BRD4
- PMID: 32968148
- PMCID: PMC7511954
- DOI: 10.1038/s41598-020-72491-9
Bardoxolone conjugation enables targeted protein degradation of BRD4
Abstract
Targeted protein degradation (TPD) has emerged as a powerful tool in drug discovery for the perturbation of protein levels using heterobifunctional small molecules. E3 ligase recruiters remain central to this process yet relatively few have been identified relative to the ~ 600 predicted human E3 ligases. While, initial recruiters have utilized non-covalent chemistry for protein binding, very recently covalent engagement to novel E3's has proven fruitful in TPD application. Herein we demonstrate efficient proteasome-mediated degradation of BRD4 by a bifunctional small molecule linking the KEAP1-Nrf2 activator bardoxolone to a BRD4 inhibitor JQ1.
Conflict of interest statement
J.A.T, J.M.M, M.S. are employees of Novartis Institutes for BioMedical Research. This study was funded by the Novartis Institutes for BioMedical Research and the Novartis-Berkeley Center for Proteomics and Chemistry Technologies. D.K.N. is a co-founder, shareholder, and adviser for Artris Therapeutic and Frontier Medicines. T.J.M., B.T., Y.X., M.L., and J.N.S. declare no competing interests.
Figures



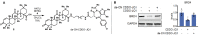
Similar articles
-
Ligandability of E3 Ligases for Targeted Protein Degradation Applications.Biochemistry. 2023 Feb 7;62(3):588-600. doi: 10.1021/acs.biochem.1c00464. Epub 2021 Sep 2. Biochemistry. 2023. PMID: 34473924 Free PMC article. Review.
-
Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications.ACS Chem Biol. 2019 Nov 15;14(11):2430-2440. doi: 10.1021/acschembio.8b01083. Epub 2019 May 13. ACS Chem Biol. 2019. PMID: 31059647 Free PMC article.
-
Discovery of a Covalent FEM1B Recruiter for Targeted Protein Degradation Applications.J Am Chem Soc. 2022 Jan 19;144(2):701-708. doi: 10.1021/jacs.1c03980. Epub 2022 Jan 7. J Am Chem Soc. 2022. PMID: 34994556 Free PMC article.
-
Alkenyl oxindole is a novel PROTAC moiety that recruits the CRL4DCAF11 E3 ubiquitin ligase complex for targeted protein degradation.PLoS Biol. 2024 May 20;22(5):e3002550. doi: 10.1371/journal.pbio.3002550. eCollection 2024 May. PLoS Biol. 2024. PMID: 38768083 Free PMC article.
-
Targeted protein degradation and the enzymology of degraders.Curr Opin Chem Biol. 2018 Jun;44:47-55. doi: 10.1016/j.cbpa.2018.05.004. Epub 2018 Jun 7. Curr Opin Chem Biol. 2018. PMID: 29885948 Review.
Cited by
-
ProxyBind: A compendium of binding sites for proximity-induced pharmacology.Comput Struct Biotechnol J. 2022 Nov 8;20:6163-6171. doi: 10.1016/j.csbj.2022.11.010. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36420167 Free PMC article.
-
Ligandability of E3 Ligases for Targeted Protein Degradation Applications.Biochemistry. 2023 Feb 7;62(3):588-600. doi: 10.1021/acs.biochem.1c00464. Epub 2021 Sep 2. Biochemistry. 2023. PMID: 34473924 Free PMC article. Review.
-
Recent Developments in PROTAC-Mediated Protein Degradation: From Bench to Clinic.Chembiochem. 2022 Jan 19;23(2):e202100270. doi: 10.1002/cbic.202100270. Epub 2021 Sep 23. Chembiochem. 2022. PMID: 34494353 Free PMC article. Review.
-
Development of MDM2 degraders based on ligands derived from Ugi reactions: Lessons and discoveries.Eur J Med Chem. 2021 Jul 5;219:113425. doi: 10.1016/j.ejmech.2021.113425. Epub 2021 Apr 4. Eur J Med Chem. 2021. PMID: 33862513 Free PMC article.
-
Reactivity of Covalent Fragments and Their Role in Fragment Based Drug Discovery.Pharmaceuticals (Basel). 2022 Nov 8;15(11):1366. doi: 10.3390/ph15111366. Pharmaceuticals (Basel). 2022. PMID: 36355538 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

