Boundary sequences flanking the mouse tyrosinase locus ensure faithful pattern of gene expression
- PMID: 32968154
- PMCID: PMC7511308
- DOI: 10.1038/s41598-020-72543-0
Boundary sequences flanking the mouse tyrosinase locus ensure faithful pattern of gene expression
Abstract
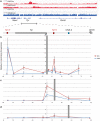
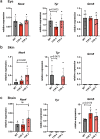
Control of gene expression is dictated by cell-type specific regulatory sequences that physically organize the structure of chromatin, including promoters, enhancers and insulators. While promoters and enhancers convey cell-type specific activating signals, insulators prevent the cross-talk of regulatory elements within adjacent loci and safeguard the specificity of action of promoters and enhancers towards their targets in a tissue specific manner. Using the mouse tyrosinase (Tyr) locus as an experimental model, a gene whose mutations are associated with albinism, we described the chromatin structure in cells at two distinct transcriptional states. Guided by chromatin structure, through the use of Chromosome Conformation Capture (3C), we identified sequences at the 5' and 3' boundaries of this mammalian gene that function as enhancers and insulators. By CRISPR/Cas9-mediated chromosomal deletion, we dissected the functions of these two regulatory elements in vivo in the mouse, at the endogenous chromosomal context, and proved their mechanistic role as genomic insulators, shielding the Tyr locus from the expression patterns of adjacent genes.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Functional dissection of the mouse tyrosinase locus control region identifies a new putative boundary activity.Nucleic Acids Res. 2003 Nov 1;31(21):6290-305. doi: 10.1093/nar/gkg793. Nucleic Acids Res. 2003. PMID: 14576318 Free PMC article.
-
The structure and function of the mouse tyrosinase locus.Pigment Cell Melanoma Res. 2021 Mar;34(2):212-221. doi: 10.1111/pcmr.12942. Epub 2020 Nov 5. Pigment Cell Melanoma Res. 2021. PMID: 33098271 Review.
-
An Efficient Vector-Based CRISPR/Cas9 System in Zebrafish Cell Line.Mar Biotechnol (NY). 2024 Jun;26(3):588-598. doi: 10.1007/s10126-024-10320-0. Epub 2024 Apr 23. Mar Biotechnol (NY). 2024. PMID: 38652190
-
Three-dimensional genome architectural CCCTC-binding factor makes choice in duplicated enhancers at Pcdhα locus.Sci China Life Sci. 2020 Jun;63(6):835-844. doi: 10.1007/s11427-019-1598-4. Epub 2020 Apr 2. Sci China Life Sci. 2020. PMID: 32249388
-
Insulators and promoters: closer than we think.Nat Rev Genet. 2010 Jun;11(6):439-46. doi: 10.1038/nrg2765. Epub 2010 May 5. Nat Rev Genet. 2010. PMID: 20442713 Free PMC article. Review.
Cited by
-
Transcription factor-mediated intestinal metaplasia and the role of a shadow enhancer.Genes Dev. 2022 Jan 1;36(1-2):38-52. doi: 10.1101/gad.348983.121. Epub 2021 Dec 30. Genes Dev. 2022. PMID: 34969824 Free PMC article.
-
Historical DNA Manipulation Overview.Methods Mol Biol. 2022;2495:3-28. doi: 10.1007/978-1-0716-2301-5_1. Methods Mol Biol. 2022. PMID: 35696025
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

