The maternal microbiome modulates fetal neurodevelopment in mice
- PMID: 32968276
- PMCID: PMC7554197
- DOI: 10.1038/s41586-020-2745-3
The maternal microbiome modulates fetal neurodevelopment in mice
Abstract
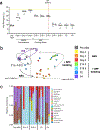
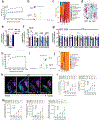
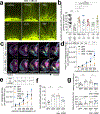
'Dysbiosis' of the maternal gut microbiome, in response to challenges such as infection1, altered diet2 and stress3 during pregnancy, has been increasingly associated with abnormalities in brain function and behaviour of the offspring4. However, it is unclear whether the maternal gut microbiome influences neurodevelopment during critical prenatal periods and in the absence of environmental challenges. Here we investigate how depletion and selective reconstitution of the maternal gut microbiome influences fetal neurodevelopment in mice. Embryos from antibiotic-treated and germ-free dams exhibited reduced brain expression of genes related to axonogenesis, deficient thalamocortical axons and impaired outgrowth of thalamic axons in response to cell-extrinsic factors. Gnotobiotic colonization of microbiome-depleted dams with a limited consortium of bacteria prevented abnormalities in fetal brain gene expression and thalamocortical axonogenesis. Metabolomic profiling revealed that the maternal microbiome regulates numerous small molecules in the maternal serum and the brains of fetal offspring. Select microbiota-dependent metabolites promoted axon outgrowth from fetal thalamic explants. Moreover, maternal supplementation with these metabolites abrogated deficiencies in fetal thalamocortical axons. Manipulation of the maternal microbiome and microbial metabolites during pregnancy yielded adult offspring with altered tactile sensitivity in two aversive somatosensory behavioural tasks, but no overt differences in many other sensorimotor behaviours. Together, our findings show that the maternal gut microbiome promotes fetal thalamocortical axonogenesis, probably through signalling by microbially modulated metabolites to neurons in the developing brain.
Conflict of interest statement
Competing Interests
Findings regarding the manipulation of the maternal microbiome to influence fetal development and sensory behavior reported in the manuscript are the subject of provisional patent application US 62/844,503, owned by UCLA. The authors declare no competing interests.
Figures










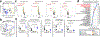
Comment in
-
Maternal microbes support fetal brain wiring.Nature. 2020 Oct;586(7828):203-205. doi: 10.1038/d41586-020-02657-y. Nature. 2020. PMID: 32968244 No abstract available.
-
Mother's microbes.Nat Rev Neurosci. 2020 Dec;21(12):666-667. doi: 10.1038/s41583-020-00401-z. Nat Rev Neurosci. 2020. PMID: 33116298 No abstract available.
References
Supplementary References:
-
- McVey Neufeld KA, Perez-Burgos A, Mao YK, Bienenstock J & Kunze WA The gut microbiome restores intrinsic and extrinsic nerve function in germ-free mice accompanied by changes in calbindin. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society 27, 627–636, doi:10.1111/nmo.12534 (2015). - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

