Fine mapping of a major locus representing the lack of prickles in eggplant revealed the availability of a 0.5-kb insertion/deletion for marker-assisted selection
- PMID: 32968346
- PMCID: PMC7495204
- DOI: 10.1270/jsbbs.20004
Fine mapping of a major locus representing the lack of prickles in eggplant revealed the availability of a 0.5-kb insertion/deletion for marker-assisted selection
Abstract
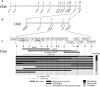
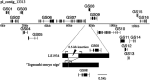
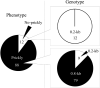
As prickles cause labour inefficiency during cultivation and scratches on the skin of fruits during transportation, they are considered undesirable traits of eggplant (Solanum melongena L.). Because the molecular basis of prickle emergence has not been entirely revealed in plants, we mapped an eggplant semi-dominant Prickle (Pl) gene locus, which causes the absence of prickles, on chromosome 6 of a linkage map of the F2 population derived from crossing the no-prickly cultivar 'Togenashi-senryo-nigo' and the prickly line LS1934. By performing synteny mapping with tomato, the genomic region corresponding to the eggplant Pl locus was identified. Through bacterial artificial chromosome (BAC) screening, positive BAC clones and the contig sequence that harbour the Pl locus in the prickly eggplant genome were revealed. The BAC contig length was 133 kb, and it contained 16 predicted genes. Among them, a characteristic 0.5-kb insertion/deletion was detected. As the 0.5-kb insertion was commonly identified with the prickly phenotype worldwide, a primer pair that amplifies the insertion/deletion could be used for marker-assisted selection of the no-prickly phenotype. Such findings contribute to map-based-cloning of the Pl gene and the understanding of gene function, ultimately providing new insights into the regulatory molecular mechanisms underlying prickle emergence in plants.
Keywords: eggplant; fine mapping; marker-assisted selection; prickle.
Copyright © 2020 by JAPANESE SOCIETY OF BREEDING.
Figures


Similar articles
-
Identification of Quantitative Trait Loci Controlling the Development of Prickles in Eggplant by Genome Re-sequencing Analysis.Front Plant Sci. 2021 Sep 8;12:731079. doi: 10.3389/fpls.2021.731079. eCollection 2021. Front Plant Sci. 2021. PMID: 34567042 Free PMC article.
-
Comparative Transcriptome Analysis Reveals Key Genes and Pathways Involved in Prickle Development in Eggplant.Genes (Basel). 2021 Feb 25;12(3):341. doi: 10.3390/genes12030341. Genes (Basel). 2021. PMID: 33668977 Free PMC article.
-
PE (Prickly Eggplant) encoding a cytokinin-activating enzyme responsible for the formation of prickles in eggplant.Hortic Res. 2024 May 10;11(7):uhae134. doi: 10.1093/hr/uhae134. eCollection 2024 Jul. Hortic Res. 2024. PMID: 38974191 Free PMC article.
-
Map-based cloning of LPD, a major gene positively regulates leaf prickle development in eggplant.Theor Appl Genet. 2024 Sep 9;137(10):216. doi: 10.1007/s00122-024-04726-6. Theor Appl Genet. 2024. PMID: 39249556
-
Detailed mapping of a resistance locus against Fusarium wilt in cultivated eggplant (Solanum melongena).Theor Appl Genet. 2016 Feb;129(2):357-67. doi: 10.1007/s00122-015-2632-8. Epub 2015 Nov 18. Theor Appl Genet. 2016. PMID: 26582508
Cited by
-
Identification of Quantitative Trait Loci Controlling the Development of Prickles in Eggplant by Genome Re-sequencing Analysis.Front Plant Sci. 2021 Sep 8;12:731079. doi: 10.3389/fpls.2021.731079. eCollection 2021. Front Plant Sci. 2021. PMID: 34567042 Free PMC article.
-
Identifying Genomic Regions Targeted During Eggplant Domestication Using Transcriptome Data.J Hered. 2021 Nov 1;112(6):519-525. doi: 10.1093/jhered/esab035. J Hered. 2021. PMID: 34130314 Free PMC article.
-
Improved genome assembly and pan-genome provide key insights into eggplant domestication and breeding.Plant J. 2021 Jul;107(2):579-596. doi: 10.1111/tpj.15313. Epub 2021 Jun 5. Plant J. 2021. PMID: 33964091 Free PMC article.
-
A Compendium for Novel Marker-Based Breeding Strategies in Eggplant.Plants (Basel). 2023 Feb 23;12(5):1016. doi: 10.3390/plants12051016. Plants (Basel). 2023. PMID: 36903876 Free PMC article. Review.
-
Comparative Transcriptome Analysis Reveals Key Genes and Pathways Involved in Prickle Development in Eggplant.Genes (Basel). 2021 Feb 25;12(3):341. doi: 10.3390/genes12030341. Genes (Basel). 2021. PMID: 33668977 Free PMC article.
References
-
- Barchi L., Lanteri S., Portis E., Stàgel A., Valè G., Toppino L. and Rotino G.L. (2010) Segregation distortion and linkage analysis in eggplant (Solanum melongena L.). Genome 53: 805–815. - PubMed
-
- Barchi L., Toppino L., Valentino D., Bassolino L., Portis E., Lanteri S. and Rotino G.L. (2018) QTL analysis reveals new eggplant loci involved in resistance to fungal wilts. Euphytica 214: 20.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

