Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy
- PMID: 32970990
- PMCID: PMC7489885
- DOI: 10.1016/j.cell.2020.09.035
Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy
Abstract
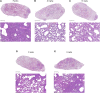
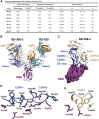
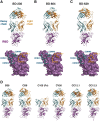
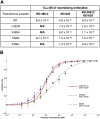
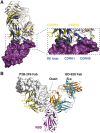
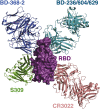
Understanding how potent neutralizing antibodies (NAbs) inhibit SARS-CoV-2 is critical for effective therapeutic development. We previously described BD-368-2, a SARS-CoV-2 NAb with high potency; however, its neutralization mechanism is largely unknown. Here, we report the 3.5-Å cryo-EM structure of BD-368-2/trimeric-spike complex, revealing that BD-368-2 fully blocks ACE2 recognition by occupying all three receptor-binding domains (RBDs) simultaneously, regardless of their "up" or "down" conformations. Also, BD-368-2 treats infected adult hamsters at low dosages and at various administering windows, in contrast to placebo hamsters that manifested severe interstitial pneumonia. Moreover, BD-368-2's epitope completely avoids the common binding site of VH3-53/VH3-66 recurrent NAbs, evidenced by tripartite co-crystal structures with RBDs. Pairing BD-368-2 with a potent recurrent NAb neutralizes SARS-CoV-2 pseudovirus at pM level and rescues mutation-induced neutralization escapes. Together, our results rationalized a new RBD epitope that leads to high neutralization potency and demonstrated BD-368-2's therapeutic potential in treating COVID-19.
Keywords: COVID-19; RBD; SARS-CoV-2; antibody cocktail; cryo-EM; epitope; hamster; mutation; neutralizing antibody; spike.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests X.S.X. and Y.C. are inventors on the patent applications of the NAbs and co-founders of Singlomics Biopharmaceuticals. Other authors declare no competing financial interests.
Figures





Similar articles
-
Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.Nature. 2020 Aug;584(7821):450-456. doi: 10.1038/s41586-020-2571-7. Epub 2020 Jul 22. Nature. 2020. PMID: 32698192
-
Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.Cell. 2020 Nov 12;183(4):1024-1042.e21. doi: 10.1016/j.cell.2020.09.037. Epub 2020 Sep 16. Cell. 2020. PMID: 32991844 Free PMC article.
-
A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.Cell. 2020 Nov 12;183(4):1058-1069.e19. doi: 10.1016/j.cell.2020.09.049. Epub 2020 Sep 23. Cell. 2020. PMID: 33058755 Free PMC article.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
-
A Structural Landscape of Neutralizing Antibodies Against SARS-CoV-2 Receptor Binding Domain.Front Immunol. 2021 Apr 28;12:647934. doi: 10.3389/fimmu.2021.647934. eCollection 2021. Front Immunol. 2021. PMID: 33995366 Free PMC article. Review.
Cited by
-
Diagnostic Value of IgM and IgG Detection in COVID-19 Diagnosis by the Mobile Laboratory B-LiFE: A Massive Testing Strategy in the Piedmont Region.Int J Environ Res Public Health. 2021 Mar 24;18(7):3372. doi: 10.3390/ijerph18073372. Int J Environ Res Public Health. 2021. PMID: 33805139 Free PMC article.
-
Prospects of animal models and their application in studies on adaptive immunity to SARS-CoV-2.Front Immunol. 2022 Sep 16;13:993754. doi: 10.3389/fimmu.2022.993754. eCollection 2022. Front Immunol. 2022. PMID: 36189203 Free PMC article. Review.
-
Computational Analysis of Mutations in the Receptor-Binding Domain of SARS-CoV-2 Spike and Their Effects on Antibody Binding.Viruses. 2022 Jan 30;14(2):295. doi: 10.3390/v14020295. Viruses. 2022. PMID: 35215888 Free PMC article.
-
Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.Nat Commun. 2022 Feb 15;13(1):871. doi: 10.1038/s41467-022-28528-w. Nat Commun. 2022. PMID: 35169135 Free PMC article.
-
Animal models for studying coronavirus infections and developing antiviral agents and vaccines.Antiviral Res. 2022 Jul;203:105345. doi: 10.1016/j.antiviral.2022.105345. Epub 2022 May 21. Antiviral Res. 2022. PMID: 35605699 Free PMC article. Review.
References
-
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. - PubMed
-
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. - PMC - PubMed
-
- Callaway E., Cyranoski D., Mallapaty S., Stoye E., Tollefson J. The coronavirus pandemic in five powerful charts. Nature. 2020;579:482–483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

