Approximating anatomically-guided PET reconstruction in image space using a convolutional neural network
- PMID: 32971267
- PMCID: PMC7812485
- DOI: 10.1016/j.neuroimage.2020.117399
Approximating anatomically-guided PET reconstruction in image space using a convolutional neural network
Abstract
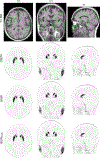
In the last two decades, it has been shown that anatomically-guided PET reconstruction can lead to improved bias-noise characteristics in brain PET imaging. However, despite promising results in simulations and first studies, anatomically-guided PET reconstructions are not yet available for use in routine clinical because of several reasons. In light of this, we investigate whether the improvements of anatomically-guided PET reconstruction methods can be achieved entirely in the image domain with a convolutional neural network (CNN). An entirely image-based CNN post-reconstruction approach has the advantage that no access to PET raw data is needed and, moreover, that the prediction times of trained CNNs are extremely fast on state of the art GPUs which will substantially facilitate the evaluation, fine-tuning and application of anatomically-guided PET reconstruction in real-world clinical settings. In this work, we demonstrate that anatomically-guided PET reconstruction using the asymmetric Bowsher prior can be well-approximated by a purely shift-invariant convolutional neural network in image space allowing the generation of anatomically-guided PET images in almost real-time. We show that by applying dedicated data augmentation techniques in the training phase, in which 16 [18F]FDG and 10 [18F]PE2I data sets were used, lead to a CNN that is robust against the used PET tracer, the noise level of the input PET images and the input MRI contrast. A detailed analysis of our CNN in 36 [18F]FDG, 18 [18F]PE2I, and 7 [18F]FET test data sets demonstrates that the image quality of our trained CNN is very close to the one of the target reconstructions in terms of regional mean recovery and regional structural similarity.
Keywords: Image reconstruction; Machine learning; Magnetic resonance imaging; Molecular imaging; Quantification.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
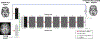
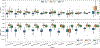



References
-
- Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viégas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, Zheng X, 2015. Tensorflow: Large-scale machine learning on heterogeneous systems, software available from tensorflow.org http://tensorflow.org/.
-
- Bowsher JE, Yuan H, Hedlung LW, Turkington TG, Akabani G, Badea A, Kurylo WC, Wheeler CT, Cofer GP, Dewhirst MW, Johnson A, 2004. Utilizing MRI information to estimate f18-FDG distributions in rat flank tumors. In: Proceedings of the Nuclear Science Symposium Conference Record IEEE 4 (C), pp. 2488–2492. doi:10.1109/NSSMIC.2004.1462760. http://ieeexplore.ieee.org/xpls/abs_all.jsp?arnumber=1462760. - DOI
-
- Burgos N, Cardoso MJ, Thielemans K, Modat M, Pedemonte S, Dickson J, Barnes A, Ahmed R, Mahoney CJ, Schott JM, Duncan JS, Atkinson D, Arridge SR, Hutton BF, Ourselin S, 2014. Attenuation correction synthesis for hybrid PET-MR scanners: application to brain studies. IEEE Trans. Med. Imaging 33 (12), 2332–2341. doi:10.1109/TMI.2014.2340135. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

