Advances in venomics: Modern separation techniques and mass spectrometry
- PMID: 32971366
- PMCID: PMC8174749
- DOI: 10.1016/j.jchromb.2020.122352
Advances in venomics: Modern separation techniques and mass spectrometry
Abstract
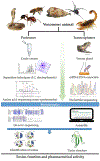
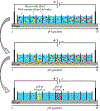
Snake venoms are complex chemical mixtures of biologically active proteins and non-protein components. Toxins have a wide range of targets and effects to include ion channels and membrane receptors, and platelet aggregation and platelet plug formation. Toxins target these effectors and effects at high affinity and selectivity. From a pharmacological perspective, snake venom compounds are a valuable resource for drug discovery and development. However, a major challenge to drug discovery using snake venoms is isolating and analyzing the bioactive proteins and peptides in these complex mixtures. Getting molecular information from complex mixtures such as snake venoms requires proteomic analyses, generally combined with transcriptomic analyses of venom glands. The present review summarizes current knowledge and highlights important recent advances in venomics with special emphasis on contemporary separation techniques and bioinformatics that have begun to elaborate the complexity of snake venoms. Several analytical techniques such as two-dimensional gel electrophoresis, RP-HPLC, size exclusion chromatography, ion exchange chromatography, MALDI-TOF-MS, and LC-ESI-QTOF-MS have been employed in this regard. The improvement of separation approaches such as multidimensional-HPLC, 2D-electrophoresis coupled to soft-ionization (MALDI and ESI) mass spectrometry has been critical to obtain an accurate picture of the startling complexity of venoms. In the case of bioinformatics, a variety of software tools such as PEAKS also has been used successfully. Such information gleaned from venomics is important to both predicting and resolving the biological activity of the active components of venoms, which in turn is key for the development of new drugs based on these venom components.
Keywords: Animal venoms; Drug discovery; Mass spectrometry; Separation methods; Venomic.
Published by Elsevier B.V.
Figures

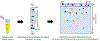

Similar articles
-
Venomous snakes of Costa Rica: biological and medical implications of their venom proteomic profiles analyzed through the strategy of snake venomics.J Proteomics. 2014 Jun 13;105:323-39. doi: 10.1016/j.jprot.2014.02.020. Epub 2014 Feb 24. J Proteomics. 2014. PMID: 24576642 Review.
-
Absolute venomics: Absolute quantification of intact venom proteins through elemental mass spectrometry.J Proteomics. 2017 Jul 5;164:33-42. doi: 10.1016/j.jprot.2017.06.001. Epub 2017 Jun 2. J Proteomics. 2017. PMID: 28579478
-
Comparison of proteomic profiles of the venoms of two of the 'Big Four' snakes of India, the Indian cobra (Naja naja) and the common krait (Bungarus caeruleus), and analyses of their toxins.Toxicon. 2017 Sep 1;135:33-42. doi: 10.1016/j.toxicon.2017.06.005. Epub 2017 Jun 8. Toxicon. 2017. PMID: 28602829
-
A Protein Decomplexation Strategy in Snake Venom Proteomics.Methods Mol Biol. 2019;1871:83-92. doi: 10.1007/978-1-4939-8814-3_5. Methods Mol Biol. 2019. PMID: 30276733
-
Venomics: unravelling the complexity of animal venoms with mass spectrometry.J Mass Spectrom. 2008 Mar;43(3):279-95. doi: 10.1002/jms.1389. J Mass Spectrom. 2008. PMID: 18302316 Review.
Cited by
-
Comparative Proteomic Analysis of the Venoms from the Most Dangerous Scorpions in Morocco: Androctonus mauritanicus and Buthus occitanus.Life (Basel). 2023 May 5;13(5):1133. doi: 10.3390/life13051133. Life (Basel). 2023. PMID: 37240778 Free PMC article.
-
The geographical distribution of scorpions, implication of venom toxins, envenomation, and potential therapeutics in Southern and Northern Africa.Toxicol Res (Camb). 2024 Aug 4;13(4):tfae118. doi: 10.1093/toxres/tfae118. eCollection 2024 Aug. Toxicol Res (Camb). 2024. PMID: 39100857 Free PMC article. Review.
-
Scorpion Venom as a Source of Antimicrobial Peptides: Overview of Biomolecule Separation, Analysis and Characterization Methods.Antibiotics (Basel). 2023 Aug 29;12(9):1380. doi: 10.3390/antibiotics12091380. Antibiotics (Basel). 2023. PMID: 37760677 Free PMC article. Review.
-
Peeking into the Stingers: A Comprehensive SWATH-MS Study of the European Hornet Vespa crabro (Linnaeus, 1758) (Hymenoptera: Vespidae) Venom Sac Extracts.Int J Mol Sci. 2024 Mar 28;25(7):3798. doi: 10.3390/ijms25073798. Int J Mol Sci. 2024. PMID: 38612607 Free PMC article.
-
Potential of Venom-Derived Compounds for the Development of New Antimicrobial Agents.Toxins (Basel). 2025 May 11;17(5):238. doi: 10.3390/toxins17050238. Toxins (Basel). 2025. PMID: 40423321 Free PMC article. Review.
References
-
- Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, Williams KL, Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it, Biotechnology & genetic engineering reviews 13 (1996) 19–50. - PubMed
-
- Jensen ON, Interpreting the protein language using proteomics, Nat Rev Mol Cell Biol 7 (2006) 391–403. - PubMed
-
- Bauw G, Van Damme J, Puype M, Vandekerckhove J, Gesser B, Ratz GP, Lauridsen JB, Celis JE, Protein-electroblotting and -microsequencing strategies in generating protein data bases from two-dimensional gels, Proceedings of the National Academy of Sciences of the United States of America, 86 (1989) 7701–7705. - PMC - PubMed
-
- Diz AP, MartÍNez-FernÁNdez M, RolÁN-Alvarez E, Proteomics in evolutionary ecology: linking the genotype with the phenotype, Molecular Ecology 21 (2012) 1060–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

