Orthohantaviruses, Emerging Zoonotic Pathogens
- PMID: 32971887
- PMCID: PMC7558059
- DOI: 10.3390/pathogens9090775
Orthohantaviruses, Emerging Zoonotic Pathogens
Abstract
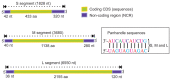
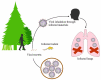
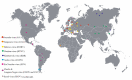
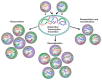
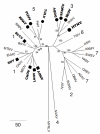
Orthohantaviruses give rise to the emerging infections such as of hemorrhagic fever with renal syndrome (HFRS) and hantavirus pulmonary syndrome (HPS) in Eurasia and the Americas, respectively. In this review we will provide a comprehensive analysis of orthohantaviruses distribution and circulation in Eurasia and address the genetic diversity and evolution of Puumala orthohantavirus (PUUV), which causes HFRS in this region. Current data indicate that the geographical location and migration of the natural hosts can lead to the orthohantaviruses genetic diversity as the rodents adapt to the new environmental conditions. The data shows that a high level of diversity characterizes the genome of orthohantaviruses, and the PUUV genome is the most divergent. The reasons for the high genome diversity are mainly caused by point mutations and reassortment, which occur in the genome segments. However, it still remains unclear whether this diversity is linked to the disease's severity. We anticipate that the information provided in this review will be useful for optimizing and developing preventive strategies of HFRS, an emerging zoonosis with potentially very high mortality rates.
Keywords: HFRS and HPS; Puumala orthohantavirus; emerging; hantavirus pulmonary syndrome; hemorrhagic fever with renal syndrome; nephropathia epidemica; reassortment; recombination; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Martinez-Valdebenito C., Calvo M., Vial C., Mansilla R., Marco C., Palma R.E., Vial P.A., Valdivieso F., Mertz G., Ferrés M. Person-to-person household and nosocomial transmission of Andes hantavirus, Southern Chile, 2011. Emerg. Infect. Dis. 2014;20:1629. doi: 10.3201/eid2010.140353. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

