Yeast Ppz1 protein phosphatase toxicity involves the alteration of multiple cellular targets
- PMID: 32973189
- PMCID: PMC7519054
- DOI: 10.1038/s41598-020-72391-y
Yeast Ppz1 protein phosphatase toxicity involves the alteration of multiple cellular targets
Abstract
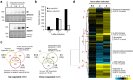
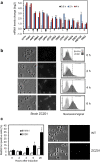
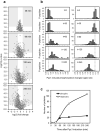
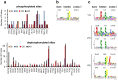
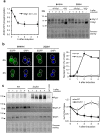
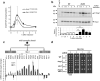
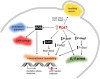
Control of the protein phosphorylation status is a major mechanism for regulation of cellular processes, and its alteration often lead to functional disorders. Ppz1, a protein phosphatase only found in fungi, is the most toxic protein when overexpressed in Saccharomyces cerevisiae. To investigate the molecular basis of this phenomenon, we carried out combined genome-wide transcriptomic and phosphoproteomic analyses. We have found that Ppz1 overexpression causes major changes in gene expression, affecting ~ 20% of the genome, together with oxidative stress and increase in total adenylate pools. Concurrently, we observe changes in the phosphorylation pattern of near 400 proteins (mainly dephosphorylated), including many proteins involved in mitotic cell cycle and bud emergence, rapid dephosphorylation of Snf1 and its downstream transcription factor Mig1, and phosphorylation of Hog1 and its downstream transcription factor Sko1. Deletion of HOG1 attenuates the growth defect of Ppz1-overexpressing cells, while that of SKO1 aggravates it. Our results demonstrate that Ppz1 overexpression has a widespread impact in the yeast cells and reveals new aspects of the regulation of the cell cycle.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Overexpression of budding yeast protein phosphatase Ppz1 impairs translation.Biochim Biophys Acta Mol Cell Res. 2020 Aug;1867(8):118727. doi: 10.1016/j.bbamcr.2020.118727. Epub 2020 Apr 24. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 32339526
-
When Phosphatases Go Mad: The Molecular Basis for Toxicity of Yeast Ppz1.Int J Mol Sci. 2022 Apr 13;23(8):4304. doi: 10.3390/ijms23084304. Int J Mol Sci. 2022. PMID: 35457140 Free PMC article. Review.
-
The toxic effects of yeast Ppz1 phosphatase are counteracted by subcellular relocalization mediated by its regulatory subunit Hal3.FEBS Lett. 2022 Jun;596(12):1556-1566. doi: 10.1002/1873-3468.14330. Epub 2022 Mar 23. FEBS Lett. 2022. PMID: 35278214
-
Saccharomyces cerevisiae protein phosphatase Ppz1 and protein kinases Sat4 and Hal5 are involved in the control of subcellular localization of Gln3 by likely regulating its phosphorylation state.J Biosci Bioeng. 2011 Mar;111(3):249-54. doi: 10.1016/j.jbiosc.2010.11.013. Epub 2011 Jan 14. J Biosci Bioeng. 2011. PMID: 21237705
-
Protein phosphatases of Saccharomyces cerevisiae.Curr Genet. 2019 Feb;65(1):41-55. doi: 10.1007/s00294-018-0884-y. Epub 2018 Sep 17. Curr Genet. 2019. PMID: 30225534 Free PMC article. Review.
Cited by
-
Functional mapping of the N-terminal region of the yeast moonlighting protein Sis2/Hal3 reveals crucial residues for Ppz1 regulation.FEBS J. 2022 Dec;289(23):7500-7518. doi: 10.1111/febs.16572. Epub 2022 Jul 18. FEBS J. 2022. PMID: 35811492 Free PMC article.
-
Comparative Analysis of Type 1 and Type Z Protein Phosphatases Reveals D615 as a Key Residue for Ppz1 Regulation.Int J Mol Sci. 2022 Jan 25;23(3):1327. doi: 10.3390/ijms23031327. Int J Mol Sci. 2022. PMID: 35163251 Free PMC article.
-
The regulatory landscape of the yeast phosphoproteome.Nat Struct Mol Biol. 2023 Nov;30(11):1761-1773. doi: 10.1038/s41594-023-01115-3. Epub 2023 Oct 16. Nat Struct Mol Biol. 2023. PMID: 37845410 Free PMC article.
-
The Toxic Effects of Ppz1 Overexpression Involve Nha1-Mediated Deregulation of K+ and H+ Homeostasis.J Fungi (Basel). 2021 Nov 25;7(12):1010. doi: 10.3390/jof7121010. J Fungi (Basel). 2021. PMID: 34946993 Free PMC article.
-
Circadian Clock Control of Translation Initiation Factor eIF2α Activity Requires eIF2γ-Dependent Recruitment of Rhythmic PPP-1 Phosphatase in Neurospora crassa.mBio. 2021 May 18;12(3):e00871-21. doi: 10.1128/mBio.00871-21. mBio. 2021. PMID: 34006661 Free PMC article.
References
-
- Posas F, Casamayor A, Morral N, Ariño J. Molecular cloning and analysis of a yeast protein phosphatase with an unusual amino-terminal region. J. Biol. Chem. 1992;267:11734–11740. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

