The CNS Myelin Proteome: Deep Profile and Persistence After Post-mortem Delay
- PMID: 32973451
- PMCID: PMC7466725
- DOI: 10.3389/fncel.2020.00239
The CNS Myelin Proteome: Deep Profile and Persistence After Post-mortem Delay
Abstract
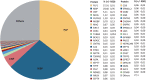
Myelin membranes are dominated by lipids while the complexity of their protein composition has long been considered to be low. However, numerous additional myelin proteins have been identified since. Here we revisit the proteome of myelin biochemically purified from the brains of healthy c56Bl/6N-mice utilizing complementary proteomic approaches for deep qualitative and quantitative coverage. By gel-free, label-free mass spectrometry, the most abundant myelin proteins PLP, MBP, CNP, and MOG constitute 38, 30, 5, and 1% of the total myelin protein, respectively. The relative abundance of myelin proteins displays a dynamic range of over four orders of magnitude, implying that PLP and MBP have overshadowed less abundant myelin constituents in initial gel-based approaches. By comparisons with published datasets we evaluate to which degree the CNS myelin proteome correlates with the mRNA and protein abundance profiles of myelin and oligodendrocytes. Notably, the myelin proteome displays only minor changes if assessed after a post-mortem delay of 6 h. These data provide the most comprehensive proteome resource of CNS myelin so far and a basis for addressing proteomic heterogeneity of myelin in mouse models and human patients with white matter disorders.
Keywords: autopsy; central nervous system (CNS); demyelination; label-free proteomics; myelin proteome; oligodendrocyte; post-mortem delay; transcriptome.
Copyright © 2020 Jahn, Siems, Kusch, Hesse, Jung, Liepold, Uecker, Sun and Werner.
Figures

Similar articles
-
Conservation and divergence of myelin proteome and oligodendrocyte transcriptome profiles between humans and mice.Elife. 2022 May 11;11:e77019. doi: 10.7554/eLife.77019. Elife. 2022. PMID: 35543322 Free PMC article.
-
Proteome Profile of Myelin in the Zebrafish Brain.Front Cell Dev Biol. 2021 Apr 8;9:640169. doi: 10.3389/fcell.2021.640169. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33898427 Free PMC article.
-
Myelin: Methods for Purification and Proteome Analysis.Methods Mol Biol. 2019;1936:37-63. doi: 10.1007/978-1-4939-9072-6_3. Methods Mol Biol. 2019. PMID: 30820892
-
Myelin proteomics: molecular anatomy of an insulating sheath.Mol Neurobiol. 2009 Aug;40(1):55-72. doi: 10.1007/s12035-009-8071-2. Epub 2009 May 19. Mol Neurobiol. 2009. PMID: 19452287 Free PMC article. Review.
-
Transgenic systems in studying myelin gene expression.Dev Neurosci. 1995;17(3):127-36. doi: 10.1159/000111281. Dev Neurosci. 1995. PMID: 8549424 Review.
Cited by
-
New Species Can Broaden Myelin Research: Suitability of Little Skate, Leucoraja erinacea.Life (Basel). 2021 Feb 11;11(2):136. doi: 10.3390/life11020136. Life (Basel). 2021. PMID: 33670172 Free PMC article.
-
The Role of Lipids, Lipid Metabolism and Ectopic Lipid Accumulation in Axon Growth, Regeneration and Repair after CNS Injury and Disease.Cells. 2021 May 1;10(5):1078. doi: 10.3390/cells10051078. Cells. 2021. PMID: 34062747 Free PMC article. Review.
-
Overview of myelin, major myelin lipids, and myelin-associated proteins.Front Chem. 2023 Feb 21;10:1041961. doi: 10.3389/fchem.2022.1041961. eCollection 2022. Front Chem. 2023. PMID: 36896314 Free PMC article. Review.
-
Shank3 mutation impairs glutamate signaling and myelination in ASD mouse model and human iPSC-derived OPCs.Sci Adv. 2024 Oct 11;10(41):eadl4573. doi: 10.1126/sciadv.adl4573. Epub 2024 Oct 11. Sci Adv. 2024. PMID: 39392881 Free PMC article.
-
Oligodendroglial fatty acid metabolism as a central nervous system energy reserve.Nat Neurosci. 2024 Oct;27(10):1934-1944. doi: 10.1038/s41593-024-01749-6. Epub 2024 Sep 9. Nat Neurosci. 2024. PMID: 39251890 Free PMC article.
References
-
- Ambrozkiewicz M. C., Schwark M., Kishimoto-Suga M., Borisova E., Hori K., Salazar-Lázaro A., et al. (2018). Polarity Acquisition in Cortical Neurons Is Driven by Synergistic Action of Sox9-Regulated Wwp1 and Wwp2 E3 Ubiquitin Ligases and Intronic miR-140. Neuron 100:1097-1115.e15. 10.1016/j.neuron.2018.10.008 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

