Description of Chloramphenicol Resistant Kineococcus rubinsiae sp. nov. Isolated From a Spacecraft Assembly Facility
- PMID: 32973710
- PMCID: PMC7472656
- DOI: 10.3389/fmicb.2020.01957
Description of Chloramphenicol Resistant Kineococcus rubinsiae sp. nov. Isolated From a Spacecraft Assembly Facility
Abstract
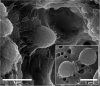
A Gram-positive, coccoid, motile, aerobic bacterium, designated strain B12T was isolated from a Jet Propulsion Laboratory spacecraft assembly cleanroom, Pasadena, CA, United States. Strain B12T was resistant to chloramphenicol (100 μg/mL), and is a relatively slow grower (3-5 days optimal). Strain B12T was found to grow optimally at 28 to 32°C, pH 7 to 8, and 0.5% NaCl. Fatty acid methyl ester analysis showed that the major fatty acid of the strain B12T was anteiso C15 : 0 (66.3%), which is also produced by other Kineococcus species. However, arachidonic acid (C20 : 4 ω6,9,12,16c) was present in strain B12T and Kineococcus glutinatus YIM 75677T but absent in all other Kineococcus species. 16S rRNA analysis revealed that strain B12T was 97.9% similar to Kineococcus radiotolerans and falls within the Kineococcus clade. Low 16S rRNA gene sequence similarities (<94%) with other genera in the family Kineosporiaceae, including Angustibacter (93%), Kineosporia (94% to 95%), Pseudokineococcus (93%), Quadrisphaera (93%), and Thalassiella (94%) demonstrated that the strain B12T does not belong to these genera. Phylogenetic analysis of the gyrB gene show that all known Kineococcus species exhibited <86% sequence similarity with B12T. Multi-locus sequence and whole genome sequence analyses confirmed that B12T clades with other Kineococcus species. Average nucleotide identity of strain B12T were 75-78% with other Kineococcus species, while values ranged from 72-75% with species from other genera within family Kineosporiaceae. Average amino-acid identities were 66-72% with other Kineococcus species, while they ranged from 50-58% with species from other genera. The dDDH comparison of strain B12T genome with members of genera Kineococcus showed 20-22% similarity, again demonstrating that B12T is distantly related to other members of the genus. Furthermore, analysis of whole proteome deduced from WGS places strain B12T in order Kineosporiales, confirming that strain B12T is a novel member of family Kineosporiaceae. Based on these analyses and other genome characteristics, strain B12T is assigned to a novel species within the genus Kineococcus, and the name Kineococcus rubinsiae sp. nov., is proposed. The type strain is B12T (=FJII-L1-CM-PAB2T; NRRL B-65556T, DSM 110506T).
Keywords: Kineococcus rubinsiae; antibiotic resistant bacteria; cleanroom; genome; spacecraft assembly facility.
Copyright © 2020 Mhatre, Singh, Wood, Parker, Pukall, Verbarg, Tindall, Neumann-Schaal and Venkateswaran.
Figures




References
-
- Chen M., Xu P., Zeng G., Yang C., Huang D., Zhang J. (2015). Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: applications, microbes and future research needs. Biotechnol. Adv. 33 745–755. 10.1016/j.biotechadv.2015.05.003 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

