The Crosstalk Between Epigenetic Mechanisms and Alternative RNA Processing Regulation
- PMID: 32973889
- PMCID: PMC7472560
- DOI: 10.3389/fgene.2020.00998
The Crosstalk Between Epigenetic Mechanisms and Alternative RNA Processing Regulation
Abstract
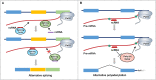
As a co-transcriptional process, RNA processing, including alternative splicing and alternative polyadenylation, is crucial for the generation of multiple mRNA isoforms. RNA processing mechanisms are widespread across all higher eukaryotes and play critical roles in cell differentiation, organ development and disease response. Recently, significant progresses have been made in understanding the mechanism of RNA processing. RNA processing is regulated by trans-acting factors such as splicing factors, RNA-binding proteins and cis-sequences in pre-mRNA, and increasing evidence suggests that epigenetic mechanisms, which are important for the dynamic regulation and state of specific chromatic regions, are also involved in co-transcriptional RNA processing. In contrast, recent studies also suggest that alternative RNA processing also has a feedback regulation on epigenetic mechanisms. In this review, we discuss recent studies and summarize the current knowledge on the epigenetic regulation of alternative RNA processing. In addition, a feedback regulation of RNA processing on epigenetic regulators is also discussed.
Keywords: DNA methylation; RNA processing; alternative polyadenylation; alternative splicing; epigenetics; histone modifications.
Copyright © 2020 Zhang, Zhang, Jiang and Duan.
Figures

Similar articles
-
Crosstalk Between mRNA 3'-End Processing and Epigenetics.Front Genet. 2021 Feb 4;12:637705. doi: 10.3389/fgene.2021.637705. eCollection 2021. Front Genet. 2021. PMID: 33613650 Free PMC article. Review.
-
Regulation of alternative splicing: Functional interplay with epigenetic modifications and its implication to cancer.Wiley Interdiscip Rev RNA. 2023 Sep 12:e1815. doi: 10.1002/wrna.1815. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37697868 Review.
-
[The epigenetic effect on pre-mRNA alternative splicing].Yi Chuan. 2014 Mar;36(3):248-55. Yi Chuan. 2014. PMID: 24846965 Review. Chinese.
-
Histone and RNA-binding protein interaction creates crosstalk network for regulation of alternative splicing.Biochem Biophys Res Commun. 2018 Apr 30;499(1):30-36. doi: 10.1016/j.bbrc.2018.03.101. Epub 2018 Mar 20. Biochem Biophys Res Commun. 2018. PMID: 29551686
-
Coupling epigenetics and RNA polyadenylation: missing links.Trends Plant Sci. 2023 Feb;28(2):223-234. doi: 10.1016/j.tplants.2022.08.023. Epub 2022 Sep 27. Trends Plant Sci. 2023. PMID: 36175275 Review.
Cited by
-
The Memory of Rice Response to Spaceflight Stress: From the Perspective of Metabolomics and Proteomics.Int J Mol Sci. 2022 Mar 21;23(6):3390. doi: 10.3390/ijms23063390. Int J Mol Sci. 2022. PMID: 35328810 Free PMC article.
-
Epigenetic Echoes: Bridging Nature, Nurture, and Healing Across Generations.Int J Mol Sci. 2025 Mar 27;26(7):3075. doi: 10.3390/ijms26073075. Int J Mol Sci. 2025. PMID: 40243774 Free PMC article. Review.
-
Position-dependent effects of RNA-binding proteins in the context of co-transcriptional splicing.NPJ Syst Biol Appl. 2023 Jan 18;9(1):1. doi: 10.1038/s41540-022-00264-3. NPJ Syst Biol Appl. 2023. PMID: 36653378 Free PMC article.
-
Genome-Wide RNAi Screening Identifies Novel Pathways/Genes Involved in Oxidative Stress and Repurposable Drugs to Preserve Cystic Fibrosis Airway Epithelial Cell Integrity.Antioxidants (Basel). 2021 Dec 2;10(12):1936. doi: 10.3390/antiox10121936. Antioxidants (Basel). 2021. PMID: 34943039 Free PMC article.
-
Intragenic heterochromatin-mediated alternative polyadenylation modulates miRNA and pollen development in rice.New Phytol. 2021 Oct;232(2):835-852. doi: 10.1111/nph.17635. Epub 2021 Aug 8. New Phytol. 2021. PMID: 34289124 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

