CXCL12-CXCR4 Interplay Facilitates Palatal Osteogenesis in Mice
- PMID: 32974338
- PMCID: PMC7471603
- DOI: 10.3389/fcell.2020.00771
CXCL12-CXCR4 Interplay Facilitates Palatal Osteogenesis in Mice
Abstract
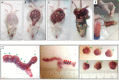
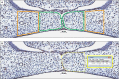
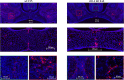
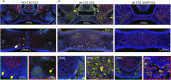
Cranial neural crest cells (CNCCs), identified by expression of transcription factor Sox9, migrate to the first branchial arch and undergo proliferation and differentiation to form the cartilage and bone structures of the orofacial region, including the palatal bone. Sox9 promotes osteogenic differentiation and stimulates CXCL12-CXCR4 chemokine-receptor signaling, which elevates alkaline phosphatase (ALP)-activity in osteoblasts to initiate bone mineralization. Disintegration of the midline epithelial seam (MES) is crucial for palatal fusion. Since we earlier demonstrated chemokine-receptor mediated signaling by the MES, we hypothesized that chemokine CXCL12 is expressed by the disintegrating MES to promote the formation of an osteogenic center by CXCR4-positive osteoblasts. Disturbed migration of CNCCs by excess oxidative and inflammatory stress is associated with increased risk of cleft lip and palate (CLP). The cytoprotective heme oxygenase (HO) enzymes are powerful guardians harnessing injurious oxidative and inflammatory stressors and enhances osteogenic ALP-activity. By contrast, abrogation of HO-1 or HO-2 expression promotes pregnancy pathologies. We postulate that Sox9, CXCR4, and HO-1 are expressed in the ALP-activity positive osteogenic regions within the CNCCs-derived palatal mesenchyme. To investigate these hypotheses, we studied expression of Sox9, CXCL12, CXCR4, and HO-1 in relation to palatal osteogenesis between E15 and E16 using (immuno)histochemical staining of coronal palatal sections in wild-type (wt) mice. In addition, the effects of abrogated HO-2 expression in HO-2 KO mice and inhibited HO-1 and HO-2 activity by administrating HO-enzyme activity inhibitor SnMP at E11 in wt mice were investigated at E15 or E16 following palatal fusion. Overexpression of Sox9, CXCL12, CXCR4, and HO-1 was detected in the ALP-activity positive osteogenic regions within the palatal mesenchyme. Overexpression of Sox9 and CXCL12 by the disintegrating MES was detected. Neither palatal fusion nor MES disintegration seemed affected by either HO-2 abrogation or inhibition of HO-activity. Sox9 progenitors seem important to maintain the CXCR4-positive osteoblast pool to drive osteogenesis. Sox9 expression may facilitate MES disintegration and palatal fusion by promoting epithelial-to-mesenchymal transformation (EMT). CXCL12 expression by the MES and the palatal mesenchyme may promote osteogenic differentiation to create osteogenic centers. This study provides novel evidence that CXCL12-CXCR4 interplay facilitates palatal osteogenesis and palatal fusion in mice.
Keywords: CXCL12-CXCR4; Sox9; cranial neural crest cells; embryology; heme oxygenase; osteogenesis; palatogenesis; pathological pregnancy.
Copyright © 2020 Verheijen, Suttorp, van Rheden, Regan, Helmich, Kuijpers-Jagtman and Wagener.
Figures









References
-
- Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B. (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16 2813–2828. 10.1101/gad.1017802 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

