SR/RS Motifs as Critical Determinants of Coronavirus Life Cycle
- PMID: 32974389
- PMCID: PMC7471607
- DOI: 10.3389/fmolb.2020.00219
SR/RS Motifs as Critical Determinants of Coronavirus Life Cycle
Abstract
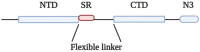
SR/RS domains are found in almost all eukaryotic genomes from C. elegans to human. These domains are thought to mediate interactions between proteins but also between proteins and RNA in complex networks associated with mRNA splicing, chromatin structure, transcription, cell cycle and cell structure. A precise and tight regulation of their function is achieved through phosphorylation of a number of serine residues within the SR/RS motifs by the Serine-Arginine protein kinases (SRPKs) that lead to delicate structural alterations. Given that coronavirus N proteins also contain SR/RS domains, we formulate the hypothesis that the viruses exploit the properties of these motifs to promote unpacking of viral RNA and virion assembly.
Keywords: N protein; SR protein kinases; SR/RS motifs; capsid assembly/disassembly; coronaviruses.
Copyright © 2020 Nikolakaki and Giannakouros.
Figures
Similar articles
-
Conserved proline-directed phosphorylation regulates SR protein conformation and splicing function.Biochem J. 2015 Mar 1;466(2):311-22. doi: 10.1042/BJ20141373. Biochem J. 2015. PMID: 25529026 Free PMC article.
-
Serine-arginine protein kinases and their targets in viral infection and their inhibition.Cell Mol Life Sci. 2023 May 17;80(6):153. doi: 10.1007/s00018-023-04808-6. Cell Mol Life Sci. 2023. PMID: 37198350 Free PMC article. Review.
-
Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif.J Mol Biol. 2016 Jun 5;428(11):2430-2445. doi: 10.1016/j.jmb.2016.04.009. Epub 2016 Apr 15. J Mol Biol. 2016. PMID: 27091468 Free PMC article.
-
Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing.Mol Cell. 1998 Apr;1(5):765-71. doi: 10.1016/s1097-2765(00)80076-3. Mol Cell. 1998. PMID: 9660960
-
Serine-arginine protein kinases: a small protein kinase family with a large cellular presence.FEBS J. 2011 Feb;278(4):570-86. doi: 10.1111/j.1742-4658.2010.07987.x. Epub 2011 Jan 12. FEBS J. 2011. PMID: 21205200 Review.
Cited by
-
Reconstitution of the SARS-CoV-2 ribonucleosome provides insights into genomic RNA packaging and regulation by phosphorylation.J Biol Chem. 2022 Nov;298(11):102560. doi: 10.1016/j.jbc.2022.102560. Epub 2022 Oct 4. J Biol Chem. 2022. PMID: 36202211 Free PMC article.
-
The variant gambit: COVID-19's next move.Cell Host Microbe. 2021 Apr 14;29(4):508-515. doi: 10.1016/j.chom.2021.02.020. Epub 2021 Mar 1. Cell Host Microbe. 2021. PMID: 33789086 Free PMC article. Review.
-
ISGylation of the SARS-CoV-2 N protein by HERC5 impedes N oligomerization and thereby viral RNA synthesis.J Virol. 2024 Sep 17;98(9):e0086924. doi: 10.1128/jvi.00869-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194248 Free PMC article.
-
First isolation of bovine coronavirus with a three-amino-acid deletion in the N gene causing severe respiratory and digestive disease in calve.Front Microbiol. 2024 Oct 1;15:1466096. doi: 10.3389/fmicb.2024.1466096. eCollection 2024. Front Microbiol. 2024. PMID: 39411436 Free PMC article.
-
Active site prediction of phosphorylated SARS-CoV-2 N-Protein using molecular simulation.Inform Med Unlocked. 2022;29:100889. doi: 10.1016/j.imu.2022.100889. Epub 2022 Feb 21. Inform Med Unlocked. 2022. PMID: 35224174 Free PMC article.
References
-
- Chang C. K., Hsu Y. L., Chang Y. H., Chao F. A., Wu M. C., Huang Y. S., et al. (2009). Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 83 2255–2264. 10.1128/JVI.02001-08 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

