Rab7D small GTPase is involved in phago-, trogocytosis and cytoskeletal reorganization in the enteric protozoan Entamoeba histolytica
- PMID: 32975360
- PMCID: PMC7757265
- DOI: 10.1111/cmi.13267
Rab7D small GTPase is involved in phago-, trogocytosis and cytoskeletal reorganization in the enteric protozoan Entamoeba histolytica
Abstract
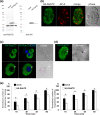
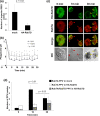
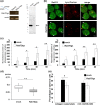
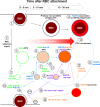
Rab small GTPases regulate membrane traffic between distinct cellular compartments of all eukaryotes in a tempo-spatially specific fashion. Rab small GTPases are also involved in the regulation of cytoskeleton and signalling. Membrane traffic and cytoskeletal regulation play pivotal role in the pathogenesis of Entamoeba histolytica, which is a protozoan parasite responsible for human amebiasis. E. histolytica is unique in that its genome encodes over 100 Rab proteins, containing multiple isotypes of conserved members (e.g., Rab7) and Entamoeba-specific subgroups (e.g., RabA, B, and X). Among them, E. histolytica Rab7 is the most diversified group consisting of nine isotypes. While it was previously demonstrated that EhRab7A and EhRab7B are involved in lysosome and phagosome biogenesis, the individual roles of other Rab7 members and their coordination remain elusive. In this study, we characterised the third member of Rab7, Rab7D, to better understand the significance of the multiplicity of Rab7 isotypes in E. histolytica. Overexpression of EhRab7D caused reduction in phagocytosis of erythrocytes, trogocytosis (meaning nibbling or chewing of a portion) of live mammalian cells, and phagosome acidification and maturation. Conversely, transcriptional gene silencing of EhRab7D gene caused opposite phenotypes in phago/trogocytosis and phagosome maturation. Furthermore, EhRab7D gene silencing caused reduction in the attachment to and the motility on the collagen-coated surface. Image analysis showed that EhRab7D was occasionally associated with lysosomes and prephagosomal vacuoles, but not with mature phagosomes and trogosomes. Finally, in silico prediction of structural organisation of EhRab7 isotypes identified unique amino acid changes on the effector binding surface of EhRab7D. Taken together, our data suggest that EhRab7D plays coordinated counteracting roles: a inhibitory role in phago/trogocytosis and lyso/phago/trogosome biogenesis, and an stimulatory role in adherence and motility, presumably via interaction with unique effectors. Finally, we propose the model in which three EhRab7 isotypes are sequentially involved in phago/trogocytosis.
Keywords: Entamoeba histolytica; Rab7D; cytoskeleton; lysosome; pathogenesis; phagocytosis; trogocytosis; vesicular traffic.
© 2020 The Authors. Cellular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Two Rab7 isotypes, EhRab7A and EhRab7B, play distinct roles in biogenesis of lysosomes and phagosomes in the enteric protozoan parasite Entamoeba histolytica.Cell Microbiol. 2007 Jul;9(7):1796-808. doi: 10.1111/j.1462-5822.2007.00915.x. Epub 2007 Mar 13. Cell Microbiol. 2007. PMID: 17359234
-
Two isotypes of phosphatidylinositol 3-phosphate-binding sorting nexins play distinct roles in trogocytosis in Entamoeba histolytica.Cell Microbiol. 2020 Mar;22(3):e13144. doi: 10.1111/cmi.13144. Epub 2019 Dec 1. Cell Microbiol. 2020. PMID: 31713312 Free PMC article.
-
Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica.J Biol Chem. 2004 Nov 19;279(47):49497-507. doi: 10.1074/jbc.M403987200. Epub 2004 Sep 3. J Biol Chem. 2004. PMID: 15347665
-
Molecular Dissection of Phagocytosis by Proteomic Analysis in Entamoeba histolytica.Genes (Basel). 2023 Jan 31;14(2):379. doi: 10.3390/genes14020379. Genes (Basel). 2023. PMID: 36833306 Free PMC article. Review.
-
Conservation and function of Rab small GTPases in Entamoeba: annotation of E. invadens Rab and its use for the understanding of Entamoeba biology.Exp Parasitol. 2010 Nov;126(3):337-47. doi: 10.1016/j.exppara.2010.04.014. Epub 2010 Apr 29. Exp Parasitol. 2010. PMID: 20434444 Review.
Cited by
-
Shaping of T Cell Functions by Trogocytosis.Cells. 2021 May 10;10(5):1155. doi: 10.3390/cells10051155. Cells. 2021. PMID: 34068819 Free PMC article. Review.
-
Entamoeba histolytica: EhADH, an Alix Protein, Participates in Several Virulence Events through Its Different Domains.Int J Mol Sci. 2024 Jul 11;25(14):7609. doi: 10.3390/ijms25147609. Int J Mol Sci. 2024. PMID: 39062867 Free PMC article.
-
De Novo Transcriptome Profiling of Naegleria fowleri Trophozoites and Cysts via RNA Sequencing.Pathogens. 2023 Jan 22;12(2):174. doi: 10.3390/pathogens12020174. Pathogens. 2023. PMID: 36839446 Free PMC article.
-
Phylogenetic reconstruction and evolution of the Rab GTPase gene family in Amoebozoa.Small GTPases. 2022 Jan;13(1):100-113. doi: 10.1080/21541248.2021.1903794. Epub 2021 Mar 29. Small GTPases. 2022. PMID: 33779495 Free PMC article.
-
The Biological Significance of Trogocytosis.Results Probl Cell Differ. 2024;73:87-129. doi: 10.1007/978-3-031-62036-2_5. Results Probl Cell Differ. 2024. PMID: 39242376 Free PMC article.
References
-
- Agarwal, S. , Anand, G. , Sharma, S. , Parimita Rath, P. , Gourinath, S. , & Bhattacharya, A. (2019). EhP3, a homolog of 14‐3‐3 family of protein participates in actin reorganization and phagocytosis in Entamoeba histolytica . PLoS Pathogens, 15(5), e1007789 10.1371/journal.ppat.1007789 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

