The requirement for cyclin E in c-Myc overexpressing breast cancers
- PMID: 32975478
- PMCID: PMC7644156
- DOI: 10.1080/15384101.2020.1804720
The requirement for cyclin E in c-Myc overexpressing breast cancers
Abstract
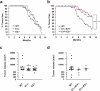
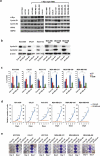
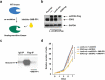
Basal-like triple-negative breast cancers frequently express high levels of c-Myc. This oncoprotein signals to the core cell cycle machinery by impinging on cyclin E. High levels of E-type cyclins (E1 and E2) are often seen in human triple-negative breast tumors. In the current study, we examined the requirement for E-type cyclins in the c-Myc-driven mouse model of breast cancer (MMTV-c-Myc mice). To do so, we crossed cyclin E1- (E1-/-) and E2- (E2-/-) deficient mice with MMTV-c-Myc animals, and observed the resulting cyclin E1-/-/MMTV-c-Myc and cyclin E2-/-/MMTV-c-Myc females for breast cancer incidence. We found that mice lacking cyclins E1 or E2 developed breast cancers like their cyclin Ewild-type counterparts. In contrast, further reduction of the dosage of E-cyclins in cyclin E1-/-E2+/-/MMTV-c-Myc and cyclin E1+/-E2-/-/MMTV-c-Myc animals significantly decreased the incidence of mammary carcinomas, revealing arole for E-cyclins in tumor initiation. We also observed that depletion of E-cyclins in human triple-negative breast cancer cell lines halted cell cycle progression, indicating that E-cyclins are essential for tumor cell proliferation. In contrast, we found that the catalytic partner of E-cyclins, the cyclin-dependent kinase 2 (CDK2), is dispensable for the proliferation of these cells. These results indicate that E-cyclins, but not CDK2, play essential and rate-limiting roles in driving the proliferation of c-Myc overexpressing breast cancer cells.
Keywords: breast cancer; c-Myc; cyclin E.
Conflict of interest statement
P.S. has been a consultant at Novartis, Genovis, Guidepoint, The Planning Shop, ORIC Pharmaceuticals, Syros and Exo Therapeutics; his laboratory receives research funding from Novartis.
Figures

Similar articles
-
Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc.Mol Cell Biol. 2009 Sep;29(17):4623-39. doi: 10.1128/MCB.00269-09. Epub 2009 Jun 29. Mol Cell Biol. 2009. PMID: 19564413 Free PMC article.
-
Kinase-independent function of E-type cyclins in liver cancer.Proc Natl Acad Sci U S A. 2018 Jan 30;115(5):1015-1020. doi: 10.1073/pnas.1711477115. Epub 2018 Jan 16. Proc Natl Acad Sci U S A. 2018. PMID: 29339491 Free PMC article.
-
Expression of cyclins E1 and E2 during mouse development and in neoplasia.Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):13138-43. doi: 10.1073/pnas.231487798. Epub 2001 Oct 30. Proc Natl Acad Sci U S A. 2001. PMID: 11687642 Free PMC article.
-
Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells.Endocr Relat Cancer. 2003 Jun;10(2):179-86. doi: 10.1677/erc.0.0100179. Endocr Relat Cancer. 2003. PMID: 12790780 Review.
-
Regulation of cell cycle entry and G1 progression by CSF-1.Mol Reprod Dev. 1997 Jan;46(1):11-8. doi: 10.1002/(SICI)1098-2795(199701)46:1<11::AID-MRD3>3.0.CO;2-U. Mol Reprod Dev. 1997. PMID: 8981358 Review.
Cited by
-
Cyclin E in normal physiology and disease states.Trends Cell Biol. 2021 Sep;31(9):732-746. doi: 10.1016/j.tcb.2021.05.001. Epub 2021 May 27. Trends Cell Biol. 2021. PMID: 34052101 Free PMC article. Review.
-
hsa_circ_0068631 promotes breast cancer progression through c-Myc by binding to EIF4A3.Mol Ther Nucleic Acids. 2021 Jul 16;26:122-134. doi: 10.1016/j.omtn.2021.07.003. eCollection 2021 Dec 3. Mol Ther Nucleic Acids. 2021. PMID: 34513299 Free PMC article.
-
Targeting cell-cycle machinery in cancer.Cancer Cell. 2021 Jun 14;39(6):759-778. doi: 10.1016/j.ccell.2021.03.010. Epub 2021 Apr 22. Cancer Cell. 2021. PMID: 33891890 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
