Expanded roles of pyruvate-sensing PdhR in transcription regulation of the Escherichia coli K-12 genome: fatty acid catabolism and cell motility
- PMID: 32975502
- PMCID: PMC7660256
- DOI: 10.1099/mgen.0.000442
Expanded roles of pyruvate-sensing PdhR in transcription regulation of the Escherichia coli K-12 genome: fatty acid catabolism and cell motility
Abstract
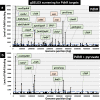
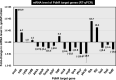
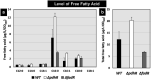
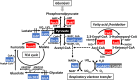
The transcription factor PdhR has been recognized as the master regulator of the pyruvate catabolism pathway in Escherichia coli, including both NAD-linked oxidative decarboxylation of pyruvate to acetyl-CoA by PDHc (pyruvate dehydrogenase complex) and respiratory electron transport of NADH to oxygen by Ndh-CyoABCD enzymes. To identify the whole set of regulatory targets under the control of pyruvate-sensing PdhR, we performed genomic SELEX (gSELEX) screening in vitro. A total of 35 PdhR-binding sites were identified along the E. coli K-12 genome, including previously identified targets. Possible involvement of PdhR in regulation of the newly identified target genes was analysed in detail by gel shift assay, RT-qPCR and Northern blot analysis. The results indicated the participation of PdhR in positive regulation of fatty acid degradation genes and negative regulation of cell mobility genes. In fact, GC analysis indicated an increase in free fatty acids in the mutant lacking PdhR. We propose that PdhR is a bifunctional global regulator for control of a total of 16-23 targets, including not only the genes involved in central carbon metabolism but also some genes for the surrounding pyruvate-sensing cellular pathways such as fatty acid degradation and flagella formation. The activity of PdhR is controlled by pyruvate, the key node between a wide variety of metabolic pathways, including generation of metabolic energy and cell building blocks.
Keywords: Escherichia coli; PdhR; cell motility; fatty acid β-oxidation; gSELEX; pyruvate; transcription regulation.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

