Periphilin self-association underpins epigenetic silencing by the HUSH complex
- PMID: 32976585
- PMCID: PMC7544229
- DOI: 10.1093/nar/gkaa785
Periphilin self-association underpins epigenetic silencing by the HUSH complex
Abstract
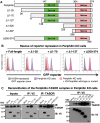
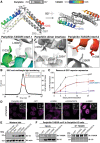
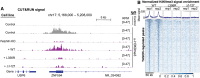
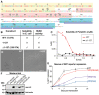
Transcription of integrated DNA from viruses or transposable elements is tightly regulated to prevent pathogenesis. The Human Silencing Hub (HUSH), composed of Periphilin, TASOR and MPP8, silences transcriptionally active viral and endogenous transgenes. HUSH recruits effectors that alter the epigenetic landscape and chromatin structure, but how HUSH recognizes target loci and represses their expression remains unclear. We identify the physicochemical properties of Periphilin necessary for HUSH assembly and silencing. A disordered N-terminal domain (NTD) and structured C-terminal domain are essential for silencing. A crystal structure of the Periphilin-TASOR minimal core complex shows Periphilin forms an α-helical homodimer, bound by a single TASOR molecule. The NTD forms insoluble aggregates through an arginine/tyrosine-rich sequence reminiscent of low-complexity regions from self-associating RNA-binding proteins. Residues required for TASOR binding and aggregation were required for HUSH-dependent silencing and genome-wide deposition of repressive mark H3K9me3. The NTD was functionally complemented by low-complexity regions from certain RNA-binding proteins and proteins that form condensates or fibrils. Our work suggests the associative properties of Periphilin promote HUSH aggregation at target loci.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Friedli M., Trono D.. The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell Dev. Biol. 2015; 31:429–451. - PubMed
-
- Zhou L., Mitra R., Atkinson P.W., Hickman A.B., Dyda F., Craig N.L.. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004; 432:995–1001. - PubMed
-
- Dupressoir A., Lavialle C., Heidmann T.. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012; 33:663–671. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 101908/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- 210688/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- BB/N011791/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MX15916/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 217191/Z/19/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

