PRosettaC: Rosetta Based Modeling of PROTAC Mediated Ternary Complexes
- PMID: 32976709
- PMCID: PMC7592117
- DOI: 10.1021/acs.jcim.0c00589
PRosettaC: Rosetta Based Modeling of PROTAC Mediated Ternary Complexes
Abstract
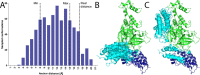
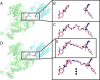
Proteolysis-targeting chimeras (PROTACs), which induce degradation by recruitment of an E3 ligase to a target protein, are gaining much interest as a new pharmacological modality. However, designing PROTACs is challenging. Formation of a ternary complex between the protein target, the PROTAC, and the recruited E3 ligase is considered paramount for successful degradation. A structural model of this ternary complex could in principle inform rational PROTAC design. Unfortunately, only a handful of structures are available for such complexes, necessitating tools for their modeling. We developed a combined protocol for the modeling of a ternary complex induced by a given PROTAC. Our protocol alternates between sampling of the protein-protein interaction space and the PROTAC molecule conformational space. Application of this protocol-PRosettaC-to a benchmark of known PROTAC ternary complexes results in near-native predictions, with often atomic accuracy prediction of the protein chains, as well as the PROTAC binding moieties. It allowed the modeling of a CRBN/BTK complex that recapitulated experimental results for a series of PROTACs. PRosettaC generated models may be used to design PROTACs for new targets, as well as improve PROTACs for existing targets, potentially cutting down time and synthesis efforts. To enable wide access to this protocol, we have made it available through a web server (https://prosettac.weizmann.ac.il/).
Conflict of interest statement
The authors declare no competing financial interest.
Figures


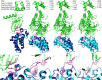


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

