Transcriptomics-Based Drug Repurposing Approach Identifies Novel Drugs against Sorafenib-Resistant Hepatocellular Carcinoma
- PMID: 32977582
- PMCID: PMC7598246
- DOI: 10.3390/cancers12102730
Transcriptomics-Based Drug Repurposing Approach Identifies Novel Drugs against Sorafenib-Resistant Hepatocellular Carcinoma
Abstract
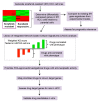
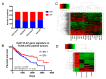
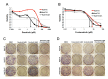
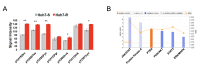
Objective: Hepatocellular carcinoma (HCC) is frequently diagnosed in patients with late-stage disease who are ineligible for curative surgical therapies. The majority of patients become resistant to sorafenib, the only approved first-line therapy for advanced cancer, underscoring the need for newer, more effective drugs. The purpose of this study is to expedite identification of novel drugs against sorafenib resistant (SR)-HCC. Methods: We employed a transcriptomics-based drug repurposing method termed connectivity mapping using gene signatures from in vitro-derived SR Huh7 HCC cells. For proof of concept validation, we focused on drugs that were FDA-approved or under clinical investigation and prioritized two anti-neoplastic agents (dasatinib and fostamatinib) with targets associated with HCC. We also prospectively validated predicted gene expression changes in drug-treated SR Huh7 cells as well as identified and validated the targets of Fostamatinib in HCC. Results: Dasatinib specifically reduced the viability of SR-HCC cells that correlated with up-regulated activity of SRC family kinases, its targets, in our SR-HCC model. However, fostamatinib was able to inhibit both parental and SR HCC cells in vitro and in xenograft models. Ingenuity pathway analysis of fostamatinib gene expression signature from LINCS predicted JAK/STAT, PI3K/AKT, ERK/MAPK pathways as potential targets of fostamatinib that were validated by Western blot analysis. Fostamatinib treatment reversed the expression of genes that were deregulated in SR HCC. Conclusion: We provide proof of concept evidence for the validity of this drug repurposing approach for SR-HCC with implications for personalized medicine.
Keywords: dasatinib; drug repurposing; drug resistance; fostamatinib; hepatocellular carcinoma; sorafenib.
Conflict of interest statement
The authors declare no conflict of interest
Figures


References
-
- Global Burden of Disease Cancer Collaboration. Fitzmaurice C., Akinyemiju T.F., Al Lami F.H., Alam T., Alizadeh-Navaei R., Allen C., Alsharif U., Alvis-Guzman N., Amini E., et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4:1553–1568. doi: 10.1001/jamaoncol.2018.2706. - DOI - PMC - PubMed
-
- Bruix J., Qin S., Merle P., Granito A., Huang Y.H., Bodoky G., Pracht M., Yokosuka O., Rosmorduc O., Breder V., et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

