Diamine Biosynthesis: Research Progress and Application Prospects
- PMID: 32978133
- PMCID: PMC7657642
- DOI: 10.1128/AEM.01972-20
Diamine Biosynthesis: Research Progress and Application Prospects
Abstract
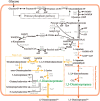
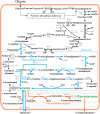
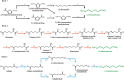
Diamines are important monomers for polyamide plastics; they include 1,3-diaminopropane, 1,4-diaminobutane, 1,5-diaminopentane, and 1,6-diaminohexane, among others. With increasing attention on environmental problems and green sustainable development, utilizing renewable raw materials for the synthesis of diamines is crucial for the establishment of a sustainable plastics industry. Recently, high-performance microbial factories, such as Escherichia coli and Corynebacterium glutamicum, have been widely used in the production of diamines. In particular, several synthetic pathways of 1,6-diaminohexane have been proposed based on glutamate or adipic acid. Here, we reviewed approaches for the biosynthesis of diamines, including metabolic engineering and biocatalysis, and the application of bio-based diamines in nylon materials. The related challenges and opportunities in the development of renewable bio-based diamines and nylon materials are also discussed.
Keywords: biosynthesis; diamines; metabolic engineering; nylon.
Copyright © 2020 American Society for Microbiology.
Figures
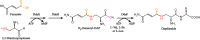
Similar articles
-
Metabolic engineering for the production of dicarboxylic acids and diamines.Metab Eng. 2020 Mar;58:2-16. doi: 10.1016/j.ymben.2019.03.005. Epub 2019 Mar 21. Metab Eng. 2020. PMID: 30905694 Review.
-
Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives.Appl Microbiol Biotechnol. 2018 Apr;102(8):3583-3594. doi: 10.1007/s00253-018-8890-z. Epub 2018 Mar 8. Appl Microbiol Biotechnol. 2018. PMID: 29520601 Review.
-
From zero to hero - production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum.Metab Eng. 2014 Sep;25:113-23. doi: 10.1016/j.ymben.2014.05.007. Epub 2014 May 14. Metab Eng. 2014. PMID: 24831706
-
Metabolic engineering of Escherichia coli for the production of 1,3-diaminopropane, a three carbon diamine.Sci Rep. 2015 Aug 11;5:13040. doi: 10.1038/srep13040. Sci Rep. 2015. PMID: 26260768 Free PMC article.
-
Metabolic engineering to improve 1,5-diaminopentane production from cellobiose using β-glucosidase-secreting Corynebacterium glutamicum.Biotechnol Bioeng. 2019 Oct;116(10):2640-2651. doi: 10.1002/bit.27082. Epub 2019 Jun 27. Biotechnol Bioeng. 2019. PMID: 31184369
Cited by
-
Degradation of 1,4-Dioxane by Xanthobacter sp. YN2.Curr Microbiol. 2021 Mar;78(3):992-1005. doi: 10.1007/s00284-021-02347-6. Epub 2021 Feb 6. Curr Microbiol. 2021. PMID: 33547937
-
Recycling and Degradation Pathways of Synthetic Textile Fibers such as Polyamide and Elastane.Glob Chall. 2025 Mar 13;9(4):2400163. doi: 10.1002/gch2.202400163. eCollection 2025 Apr. Glob Chall. 2025. PMID: 40255241 Free PMC article. Review.
-
Improving growth properties of Corynebacterium glutamicum by implementing an iron-responsive protocatechuate biosynthesis.Microb Biotechnol. 2023 May;16(5):1041-1053. doi: 10.1111/1751-7915.14244. Epub 2023 Mar 11. Microb Biotechnol. 2023. PMID: 36905370 Free PMC article.
-
Metagenomics and metabolomics analysis to investigate the effect of Shugan decoction on intestinal microbiota in irritable bowel syndrome rats.Front Microbiol. 2022 Nov 21;13:1024822. doi: 10.3389/fmicb.2022.1024822. eCollection 2022. Front Microbiol. 2022. PMID: 36478867 Free PMC article.
-
Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum.Bioengineering (Basel). 2023 Jan 24;10(2):157. doi: 10.3390/bioengineering10020157. Bioengineering (Basel). 2023. PMID: 36829651 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

