The Household Resistome: Frequency of β-Lactamases, Class 1 Integrons, and Antibiotic-Resistant Bacteria in the Domestic Environment and Their Reduction during Automated Dishwashing and Laundering
- PMID: 32978137
- PMCID: PMC7657631
- DOI: 10.1128/AEM.02062-20
The Household Resistome: Frequency of β-Lactamases, Class 1 Integrons, and Antibiotic-Resistant Bacteria in the Domestic Environment and Their Reduction during Automated Dishwashing and Laundering
Abstract
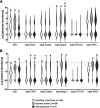
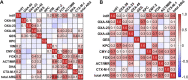
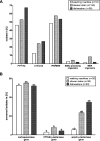
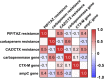
Households provide a habitat for bacteria originating from humans, animals, foods, contaminated clothes, or other sources. Thus, bacteria carrying antibiotic resistance genes (ARGs) may be introduced via household members, animals, or the water supply from external habitats into private households and vice versa. Since data on antibiotic resistance (ABR) in the domestic environment are limited, this study aimed to determine the abundance of β-lactamase, mobile colistin resistance, and class 1 integron genes and the correlation of their presence and to characterize phenotypically resistant strains in 54 private households in Germany. Additionally, the persistence of antibiotic-resistant bacteria during automated dishwashing compared to that during laundering was assessed. Shower drains, washing machines, and dishwashers were sampled and analyzed using quantitative real-time PCR. Resistant strains were isolated, followed by identification and antibiotic susceptibility testing using a Vitek 2 system. The results showed a significantly higher relative ARG abundance of 0.2367 ARG copies/16S rRNA gene copies in shower drains than in dishwashers (0.1329 ARG copies/16S rRNA gene copies) and washing machines (0.0006 ARG copies/16S rRNA gene copies). blaCMY-2, blaACT/MIR, and blaOXA-48 were the most prevalent ARG, and intI1 occurred in 96.3% of the households, while no mcr genes were detected. Several β-lactamase genes co-occurred, and the resistance of bacterial isolates correlated positively with genotypic resistance, with carbapenemase genes dominating across isolates. Antibiotic-resistant bacteria were significantly reduced during automated dishwashing as well as laundering tests and did not differ from susceptible strains. Overall, the domestic environment may represent a potential reservoir of β-lactamase genes and β-lactam-resistant bacteria, with shower drains being the dominant source of ABR.IMPORTANCE The abundance of antibiotic-resistant bacteria and ARGs is steadily increasing and has been comprehensively analyzed in natural environments, animals, foods, and wastewater treatment plants. In this respect, β-lactams and colistin are of particular interest due to the emergence of multidrug-resistant Gram-negative bacteria. Despite the connection of private households to these environments, only a few studies have focused on the domestic environment so far. Therefore, the present study further investigated the occurrence of ARGs and antibiotic-resistant bacteria in shower drains, washing machines, and dishwashers. The analysis of the domestic environment as a potential reservoir of resistant bacteria is crucial to determine whether households contribute to the spread of ABR or may be a habitat where resistant bacteria from the natural environment, humans, food, or water are selected due to the use of detergents, antimicrobial products, and antibiotics. Furthermore, ABR could limit the options for the treatment of infections arising in the domestic environment.
Keywords: antibiotic resistance; class 1 integron; domestic environment; household; multiresistance; β-lactamases.
Copyright © 2020 Schages et al.
Figures

Similar articles
-
Prevalence of β-lactamase genes in domestic washing machines and dishwashers and the impact of laundering processes on antibiotic-resistant bacteria.J Appl Microbiol. 2017 Dec;123(6):1396-1406. doi: 10.1111/jam.13574. Epub 2017 Oct 13. J Appl Microbiol. 2017. PMID: 28845592
-
Integron-mediated multidrug resistance in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from fecal specimens in Egypt.J Egypt Public Health Assoc. 2016 Jun;91(2):73-9. doi: 10.1097/01.EPX.0000483165.56114.d8. J Egypt Public Health Assoc. 2016. PMID: 27455084
-
Occurrence of IMP-8, IMP-10, and IMP-13 metallo-β-lactamases located on class 1 integrons and other extended-spectrum β-lactamases in bacterial isolates from Tunisian rivers.Scand J Infect Dis. 2013 Feb;45(2):95-103. doi: 10.3109/00365548.2012.717712. Epub 2012 Sep 19. Scand J Infect Dis. 2013. PMID: 22992193
-
The role of the natural aquatic environment in the dissemination of extended spectrum beta-lactamase and carbapenemase encoding genes: A scoping review.Water Res. 2020 Aug 1;180:115880. doi: 10.1016/j.watres.2020.115880. Epub 2020 May 7. Water Res. 2020. PMID: 32438141
-
Plasmid-mediated resistance is going wild.Plasmid. 2018 Sep;99:99-111. doi: 10.1016/j.plasmid.2018.09.010. Epub 2018 Sep 21. Plasmid. 2018. PMID: 30243983 Review.
Cited by
-
Assessing Antibiotic-Resistant Genes in University Dormitory Washing Machines.Microorganisms. 2024 May 30;12(6):1112. doi: 10.3390/microorganisms12061112. Microorganisms. 2024. PMID: 38930496 Free PMC article.
-
Bacterial Contamination in the Different Parts of Household Washing Machine: New Insights from Chengdu, Western China.Curr Microbiol. 2024 Mar 13;81(5):114. doi: 10.1007/s00284-024-03630-y. Curr Microbiol. 2024. PMID: 38478167
-
Does Antibiotic Use Contribute to Biofilm Resistance in Sink Drains? A Case Study from Four German Hospital Wards.Antibiotics (Basel). 2024 Dec 1;13(12):1148. doi: 10.3390/antibiotics13121148. Antibiotics (Basel). 2024. PMID: 39766538 Free PMC article.
-
Biological and Synthetic Surfactants Increase Class I Integron Prevalence in Ex Situ Biofilms.Microorganisms. 2024 Mar 31;12(4):712. doi: 10.3390/microorganisms12040712. Microorganisms. 2024. PMID: 38674656 Free PMC article.
-
Virucidal Efficacy of Laundering.Pathogens. 2022 Aug 31;11(9):993. doi: 10.3390/pathogens11090993. Pathogens. 2022. PMID: 36145425 Free PMC article. Review.
References
-
- WHO. 2019. Critically important antimicrobials for human medicine, 6th revision WHO, Geneva, Switzerland.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

