Development and validation of a high-throughput whole cell assay to investigate Staphylococcus aureus adhesion to host ligands
- PMID: 32978256
- PMCID: PMC7864066
- DOI: 10.1074/jbc.RA120.015360
Development and validation of a high-throughput whole cell assay to investigate Staphylococcus aureus adhesion to host ligands
Abstract
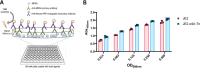
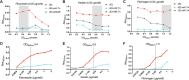
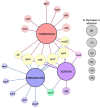
Staphylococcus aureus adhesion to the host's skin and mucosae enables asymptomatic colonization and the establishment of infection. This process is facilitated by cell wall-anchored adhesins that bind to host ligands. Therapeutics targeting this process could provide significant clinical benefits; however, the development of anti-adhesives requires an in-depth knowledge of adhesion-associated factors and an assay amenable to high-throughput applications. Here, we describe the development of a sensitive and robust whole cell assay to enable the large-scale profiling of S. aureus adhesion to host ligands. To validate the assay, and to gain insight into cellular factors contributing to adhesion, we profiled a sequence-defined S. aureus transposon mutant library, identifying mutants with attenuated adhesion to human-derived fibronectin, keratin, and fibrinogen. Our screening approach was validated by the identification of known adhesion-related proteins, such as the housekeeping sortase responsible for covalently linking adhesins to the cell wall. In addition, we also identified genetic loci that could represent undescribed anti-adhesive targets. To compare and contrast the genetic requirements of adhesion to each host ligand, we generated a S. aureus Genetic Adhesion Network, which identified a core gene set involved in adhesion to all three host ligands, and unique genetic signatures. In summary, this assay will enable high-throughput chemical screens to identify anti-adhesives and our findings provide insight into the target space of such an approach.
Keywords: MRSA; Staphylococcus aureus; anti-adhesives; antibiotic resistance; antivirulence; bacterial adhesion; cell wall-anchored proteins; drug discovery; high-throughput screening; methicillin-resistant Staphylococcus aureus (MRSA); virulence factor.
© 2020 Petrie et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Johnny W.P. Bacterial pathogenesis. In: Barron S., editor. Medical Microbiology. 4 Ed. 1996. Galveston, TX.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

