SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response
- PMID: 32979316
- PMCID: PMC7500949
- DOI: 10.1016/j.stem.2020.09.013
SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response
Abstract
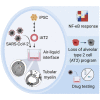
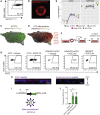
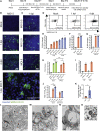
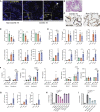
A hallmark of severe COVID-19 pneumonia is SARS-CoV-2 infection of the facultative progenitors of lung alveoli, the alveolar epithelial type 2 cells (AT2s). However, inability to access these cells from patients, particularly at early stages of disease, limits an understanding of disease inception. Here, we present an in vitro human model that simulates the initial apical infection of alveolar epithelium with SARS-CoV-2 by using induced pluripotent stem cell-derived AT2s that have been adapted to air-liquid interface culture. We find a rapid transcriptomic change in infected cells, characterized by a shift to an inflammatory phenotype with upregulation of NF-κB signaling and loss of the mature alveolar program. Drug testing confirms the efficacy of remdesivir as well as TMPRSS2 protease inhibition, validating a putative mechanism used for viral entry in alveolar cells. Our model system reveals cell-intrinsic responses of a key lung target cell to SARS-CoV-2 infection and should facilitate drug development.
Keywords: COVID-19; SARS-CoV-2; alveolar epithelial cell; alveolar type 2 cell; human induced pluripotent stem cells; iPSCs; inflammation; lung.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

Update of
-
SARS-CoV-2 Infection of Pluripotent Stem Cell-derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response.bioRxiv [Preprint]. 2020 Aug 6:2020.06.30.175695. doi: 10.1101/2020.06.30.175695. bioRxiv. 2020. Update in: Cell Stem Cell. 2020 Dec 3;27(6):962-973.e7. doi: 10.1016/j.stem.2020.09.013. PMID: 32637964 Free PMC article. Updated. Preprint.
References
-
- Abo K.M., Ma L., Matte T., Huang J., Alysandratos K.D., Werder R.B., Mithal A., Beermann M.L., Lindstrom-Vautrin J., Mostoslavsky G. Human iPSC-derived alveolar and airway epithelial cells can be cultured at air-liquid interface and express SARS-CoV-2 host factors. bioRxiv. 2020 2020.2006.2003.132639.
-
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI135912/AI/NIAID NIH HHS/United States
- U01 TR001810/TR/NCATS NIH HHS/United States
- T32 HL007035/HL/NHLBI NIH HHS/United States
- U01 HL134745/HL/NHLBI NIH HHS/United States
- R01 HL139799/HL/NHLBI NIH HHS/United States
- R01 DK117940/DK/NIDDK NIH HHS/United States
- R01 HL095993/HL/NHLBI NIH HHS/United States
- UL1 TR001430/TR/NCATS NIH HHS/United States
- F30 HL147426/HL/NHLBI NIH HHS/United States
- R01 DK101501/DK/NIDDK NIH HHS/United States
- U01 HL134766/HL/NHLBI NIH HHS/United States
- U01 HL148692/HL/NHLBI NIH HHS/United States
- 75N92020C00005/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

