Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection
- PMID: 32979593
- PMCID: PMC7502227
- DOI: 10.1016/j.bios.2020.112642
Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection
Abstract
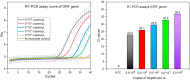
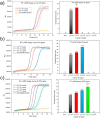
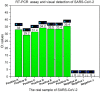
The outbreaks of the infectious disease COVID-19 caused by SARS-CoV-2 seriously threatened the life of humans. A rapid, reliable and specific detection method was urgently needed. Herein, we reported a contamination-free visual detection method for SARS-CoV-2 with LAMP and CRISPR/Cas12a technology. CRISPR/Cas12a reagents were pre-added on the inner wall of the tube lid. After LAMP reaction, CRISPR/Cas12a reagents were flowed into the tube and mixed with amplicon solution by hand shaking, which can effectively avoid possible amplicon formed aerosol contamination caused by re-opening the lid after amplification. CRISPR/Cas12a can highly specific recognize target sequence and discriminately cleave single strand DNA probes (5'-6FAM 3'-BHQ1). With smart phone and portable 3D printing instrument, the produced fluorescence can be seen by naked eyes without any dedicated instruments, which is promising in the point-of-care detection. The whole amplification and detection process could be completed within 40 min with high sensitivity of 20 copies RNA of SARS-CoV-2. This reaction had high specificity and could avoid cross-reactivity with other common viruses such as influenza virus. For 7 positive and 3 negative respiratory swab samples provided by Zhejiang Provincial Center for Disease Control and Prevention, our detection results had 100% positive agreement and 100% negative agreement, which demonstrated the accuracy and application prospect of this method.
Keywords: CRISPR/Cas12a; RT-LAMP; SARS-CoV-2; Visual detection.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

