Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13)
- PMID: 32980406
- PMCID: PMC7513821
- DOI: 10.1016/j.ijbiomac.2020.09.138
Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13)
Abstract
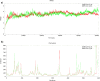
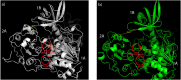
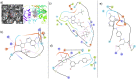
SARS-CoV-2 has caused COVID-19 outbreak with nearly 2 M infected people and over 100K death worldwide, until middle of April 2020. There is no confirmed drug for the treatment of COVID-19 yet. As the disease spread fast and threaten human life, repositioning of FDA approved drugs may provide fast options for treatment. In this aspect, structure-based drug design could be applied as a powerful approach in distinguishing the viral drug target regions from the host. Evaluation of variations in SARS-CoV-2 genome may ease finding specific drug targets in the viral genome. In this study, 3458 SARS-CoV-2 genome sequences isolated from all around the world were analyzed. Incidence of C17747T and A17858G mutations were observed to be much higher than others and they were on Nsp13, a vital enzyme of SARS-CoV-2. Effect of these mutations was evaluated on protein-drug interactions using in silico methods. The most potent drugs were found to interact with the key and neighbor residues of the active site responsible from ATP hydrolysis. As result, cangrelor, fludarabine, folic acid and polydatin were determined to be the most potent drugs which have potency to inhibit both the wild type and mutant SARS-CoV-2 helicase. Clinical data supporting these findings would be important towards overcoming COVID-19.
Keywords: Drug repositioning; Helicase; Mutation analysis; Nsp13; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Figures




References
-
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. - DOI - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

